library(dplyr) # for data wrangling
library(modelsummary) # for summary tables
options(modelsummary_factory_latex = "kableExtra")
options(modelsummary_factory_html = "kableExtra")6 Multicollinearity
Learning objectives
- Be able to describe and identify different forms of multicollinearity.
- Understand the effects of multicollinearity on parameter estimates and their standard errors.
- Be able to evaluate predictors for multicollinearity using variance inflation factors.
- Understand some strategies for dealing with multicollinearity.
6.1 R Packages
We begin by loading the dplyr package:
In addition, we will use data and functions from the following packages:
openintrofor themammalsdata setcarfor theviffunction used to calculate variance inflation factorsGGallyfor creating a scatterplot matrix
6.2 Multicollinearity
As we noted in Section 3.14.1, predictor variables will almost always be correlated in observational studies. When these correlations are strong, it can be difficult to estimate parameters in a multiple regression model, leading to coefficients with large standard errors. This should, perhaps, not be too surprising given that we have emphasized the interpretation of regression coefficients in a multiple regression model as quantifying the expected change in
If we are primarily interested in prediction, collinearity may be of little importance. We can use a model with collinear variables to obtain predictions, and these predictions can be unbiased and precise as long as our data are representative of the population for which we aim to obtain predictions (Michael H. Kutner et al. 2005, 5:283; Dormann et al. 2013). On the other hand, collinearity will make inference challenging since it will be difficult to distinguish the unique effects of collinear variables. Thus, we will need tools to identify collinear variables and to make inferences regarding their effects.
Ecologists commonly compute pairwise correlations between all predictor variables and drop one member of any pair when the absolute value of the associated correlation coefficient,
There are several types of collinearity/multicollinearity that can arise when analyzing data. In some cases, we may have multiple measurements that reflect some latent (i.e. unobserved) quantity; Dormann et al. (2013) refers to this as intrinsic collinearity. For example, biologists may quantify the size of their study animals using multiple measurements (e.g., length from head to tail, neck circumference, etc). It would not be surprising to find that pairwise correlations between these different body measurements are large because they all relate to one latent quantity, which is overall body size. In other cases, quantitative variables may be constructed by aggregating categorical variables (e.g., percent cover associated with different land-use categories in different pixels within a Geographical Information System [GIS] layer). These compositional variables must sum to 1, and thus, the last category is completely determined by the others. This is an extreme example of multicollinearity; statistical software will be unable to estimate separate coefficients for each compositional variable along with an overall intercept. For a discussion of collinearity in the context of compositional variables and potential solutions, see Valle, Mintz, and Brack (2024).
Collinearity can also occur structurally, e.g., when fitting models with polynomial terms or with predictors that have very different scales. For example, the correlation between
x <- seq(0:100); x2 = x*x
cor(x,x^2)[1] 0.9688484This type of collinearity can often be fixed by using various transformations of the explanatory variables e.g., using orthogonal polynomials as discussed in 4.11. Lastly, variables may be correlated due to other reasons that are harder to identify on their own - e.g., perhaps several predictors covary spatially leading to mild to severe multicollinearity. As an example, temperature, precipitation, and elevation may all covary along an elevation gradient. Lastly, collinearity can also happen by chance in small data sets; Dormann et al. (2013) calls this incidental collinearity and suggests it is best addressed through proper sampling or experimental design (e.g., to ensure that predictor variables do not substantially covary).
Some of the symptoms of multicollinearity include:
- regression coefficients may be statistically significant in simple linear regression models but not in regression models with multiple predictors as they compete to explain the same variability in
- parameter estimates may have large standard errors, representing large uncertainty, despite large sample sizes.
- the p-values associated with t-tests for the individual coefficients in the regression model may be large (
- lack of convergence when using numerical optimization routines to estimate parameters in complex statistical models.
- parameter estimates may be unstable; small changes in the data or model structure can change parameter estimates considerably (sometimes referred to as the “bouncing beta problem”).
- parameter estimates may change in magnitude (and even sign) depending on what other variables are included in the model (e.g., Table 6.1). As we will see in Chapter 7, we can use causal diagrams to try to understand and leverage these types of behaviors when estimating direct and indirect effects of our explanatory variables.
6.3 Motivating example: what factors predict how long mammals sleep?
To demonstrate some of these issues, let’s briefly explore a popular data set from the openintro package (Çetinkaya-Rundel et al. 2021) containing measurements of the average amount of time different mammals sleep per 24 hour period (Allison and Cicchetti 1976; Savage and West 2007).
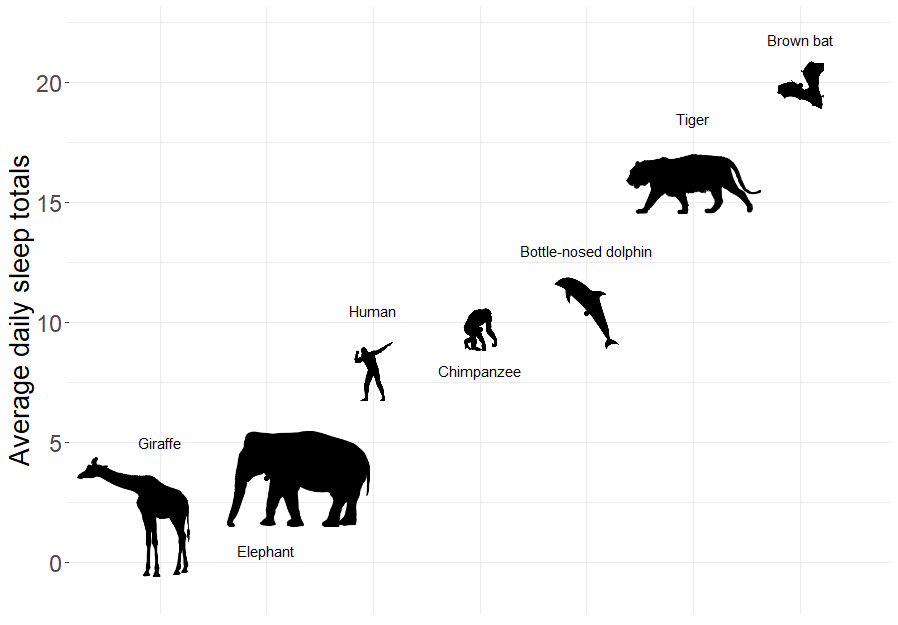
Sleep is characterized by either slow wave (non-dreaming) or rapid eye movement (dreaming), with wide variability in the amount of both types of sleep among mammals. In particular, Roe Deer (Capreolus capreolus) sleep < 3 hours/day whereas the Little Brown Bat (Myotis lucifugus) sleeps close to 20 hours per day.
The data set has many possible variables we could try to use to predict the amount of time different mammals sleep, including:
- Lifespan (years)
- Gestation (days)
- Brain weight (g)
- Body weight (kg)
- Predation Index (1-5; 1 = least likely to be preyed upon)
- Exposure Index [1-5: 1 = least exposed (e.g., animal sleeps in a den)]
- Danger Index (1:5, combines exposure and predation; 1= least danger from other animals)
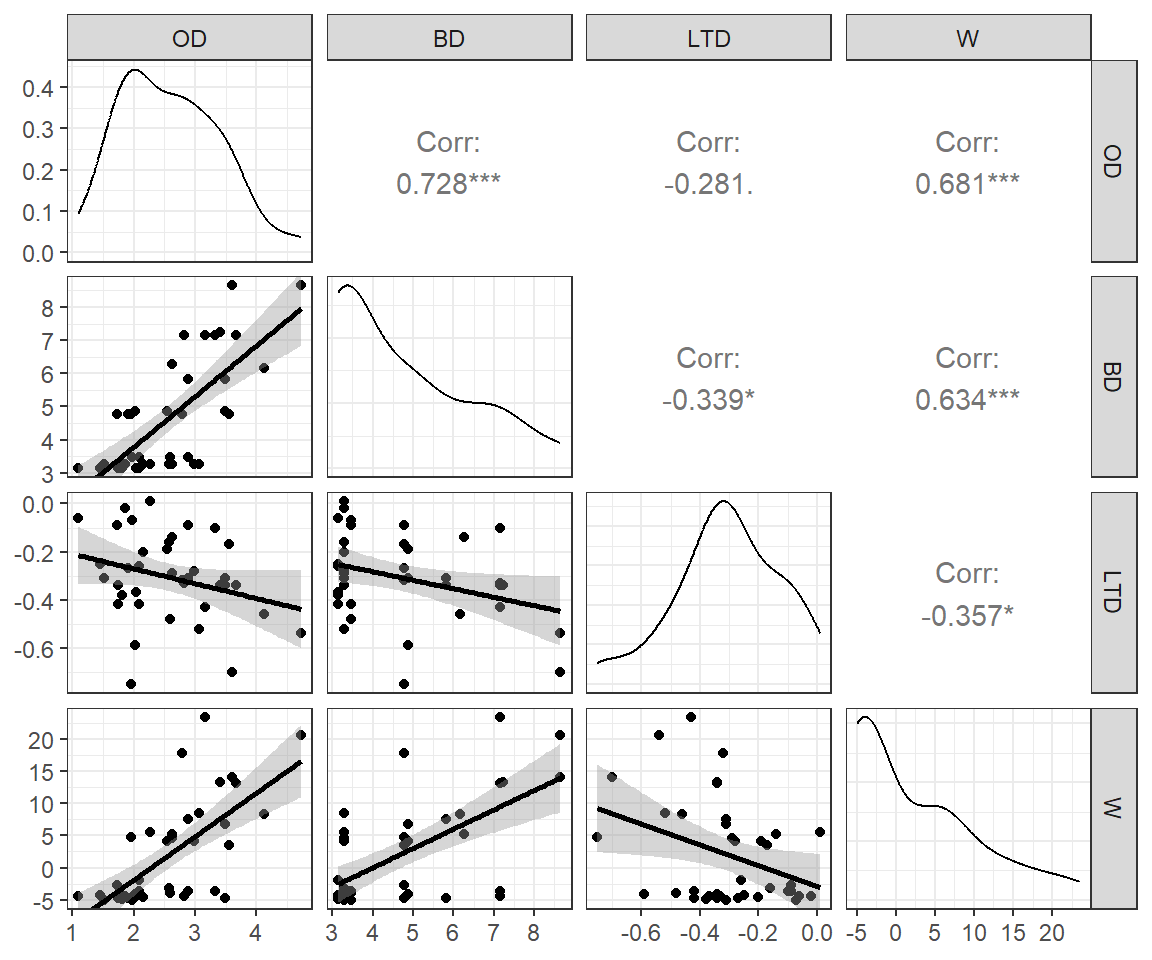
It turns out that all of these variables are negatively correlated with sleep (see the correlations in the first row of Figure 6.2) - no wonder it is so difficult to get a good night’s rest!
library(openintro)
data(mammals, package="openintro")
mammals <- mammals %>% dplyr::select(total_sleep, life_span, gestation,
brain_wt, body_wt, predation,
exposure, danger) %>%
filter(complete.cases(.))
GGally::ggpairs(mammals)
mammals data set.
We also see that our predictor variables are highly correlated with each other (Figure 6.2). Lastly, we see that the distribution of brain_wt and body_wt are severely right skewed with a few large outliers. To reduce the impact of these outliers, we will consider log-transformed predictors, log(brain_wt) and log(body_wt) when including them in regression models1.
To see how collinearity can impact estimated regression coefficients and statistical hypothesis tests involving these coefficients, let’s consider the following two models:
Model 1: Sleep = life_span
Model 2: Sleep = life_span danger log(brain_wt)
model2<-lm(total_sleep ~ life_span + danger + log(brain_wt), data=mammals)
modelsummary(list("Model 1" = model1, "Model 2" = model2), gof_omit = ".*",
estimate = "{estimate} ({p.value})",
statistic = NULL)In the first model, life_span is statistically significant and has a coefficient of -0.097 (Table 6.1). However, life_span is no longer statistically significant once we include danger and log(brain_wt), and its coefficient is also an order of magnitude smaller (
Because life_span and log(brain_wt) are positively correlated, they will “compete” to explain the same variability in sleep measurements.
mosaic::cor(life_span ~ log(brain_wt), data = mammals, use = "complete.obs")[1] 0.7267191If we try to fit a model with all of the explanatory variables, we find that only danger and predation have p-values < 0.05 (Table 6.2). Further, the coefficient for predation is positive despite our intuition that high predation pressure should be negatively correlated with sleep. And, indeed, the coefficient for predation is negative if it is the only explanatory variable in the model. In Section 6.5 and Chapter 7, we will use causal diagrams to explore potential reasons why the magnitude and direction of a regression coefficient may change after adding or excluding a variable.
model3 <- lm(total_sleep ~ life_span + gestation + log(brain_wt) +
log(body_wt) + predation + exposure + danger, data=mammals)
model4 <- lm(total_sleep ~ predation, data=mammals)
modelsummary(list("Model 3" = model3, "Model 4" = model4), gof_omit = ".*",
estimate = "{estimate} ({p.value})",
statistic = NULL)| Model 3 | Model 4 | |
|---|---|---|
| (Intercept) | 17.149 (<0.001) | 14.465 (<0.001) |
| life_span | 0.015 (0.647) | |
| gestation | -0.008 (0.094) | |
| log(brain_wt) | -0.841 (0.189) | |
| log(body_wt) | 0.129 (0.781) | |
| predation | 1.950 (0.046) | -1.448 (<0.001) |
| exposure | 0.828 (0.147) | |
| danger | -4.193 (<0.001) |
6.4 Variance inflation factors (VIF)
Variance inflation factors (VIFs) were developed to quantify collinearity in ordinary least squares regression models. They measure the degree to which the variance (i.e., SE
where
Let’s have a look at the VIFs for the sleep data if we were to include all of the predictors. We will use the vif function in the car package (Fox and Weisberg 2019) to calculate the VIFs:
car::vif(model3) life_span gestation log(brain_wt) log(body_wt) predation
2.507122 2.971837 16.568907 13.903451 12.978738
exposure danger
5.121031 16.732626 We can also use the check_model function in the performance package to create a nice visualization of the VIFS (Figure 6.3). We see that several of the VIFs are large and greater than 10, suggesting that several of our predictor variables are collinear. You will have a chance to further explore this data set as part of an exercise associated with this Section.
performance::check_model(model3, check = "vif")
check_model function in the performance package (Lüdecke et al. 2021).
6.5 Understanding confounding using DAGs: A simulation example
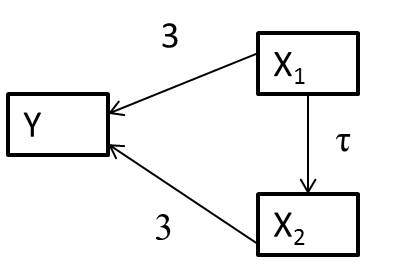
Let’s consider a system of variables, represented using a directed acylical graph (DAG)2 in which arrows represent causal effects (Figure 6.4).
The arrows between
Let’s simulate data consistent with the DAG in Figure 6.4, assuming:
Let’s simulate data sets for values of
lm(Y ~ X1)lm(Y ~ X1 + X2)
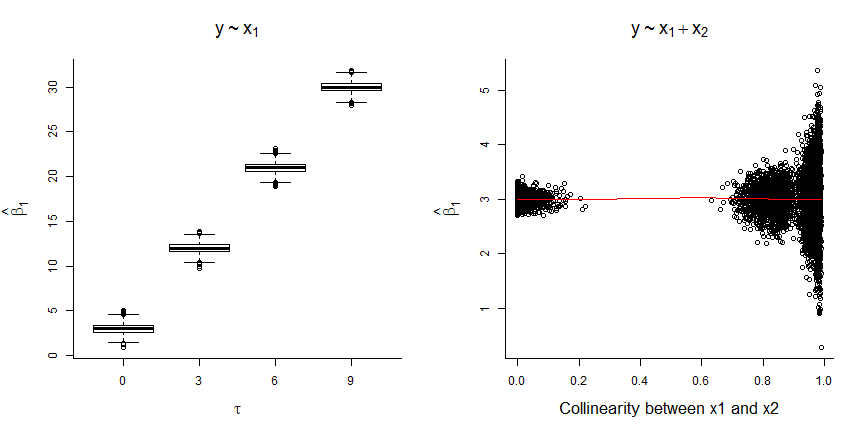
Looking at Figure 6.5, we see that:
- The coefficient for
- The magnitude of the bias increases with the correlation between
- The coefficient for
To understand these results, note that:
Thus, when we leave
6.6 Strategies for addressing multicollinearity
As we saw in the last section, omitting important predictors can lead to biased regression parameter estimators.3 Yet, it is common for researchers to drop one or more variables when they are highly correlated. For example, when faced with 2 highly correlated predictors, users may compare univariate regression models and select the variable that has the strongest association with the response variable. Alternatively, if one applies a stepwise model-selection routine (Section 8.4), it is likely that one of the highly correlated predictors will be dropped during that process because competing predictors will often look “underwhelming” when they are both included. What should we do instead?
As we will discuss in Chapter 8, it is paramount to consider how a model will be used when developing strategies for handling messy data situations, including multicollinearity. If our goal is to generate accurate predictions, then we may not care about the effect of any one variable in isolation (i.e., we may not be interested in how
Alternatively, one may create new predictor variables that are orthogonal (i.e., not correlated), but these methods present their own challenges when it comes to interpretation. In the next sections, we will consider two such approaches suggested by Graham (2003):
- Residual and sequential regression
- Principal components regression
Graham (2003) also includes a short overview of structural equation models, which have a rich history, particularly in the social sciences and deserve their own treatment (by someone more well versed in their use than me!). One option would be Jarrett Byrnes’s course. For a nice introduction, I also suggest reading Grace (2008).
6.8 Residual and sequential regression
Graham (2003) initially considers an approach he refers to as residual and sequential regression, in which variables are prioritized in terms of their order of entry into the model, with higher-priority variables allowed to account for unique as well as shared contributions to explaining the variability of the response variable,
Consider a situation in which you are interested in building a model that includes three correlated predictor variables. To apply this approach, we begin by ordering the variables based on their a priori importance, say
In this case,
For the Kelp data set, Graham (2003) decided on the following order of greatest to least importance, wave orbital displacement (OD), wind velocity (W), average tidal height (LTD), and then wave breaking depth (BD). He then created the following predictors:
Kelp$W.g.OD<-lm(W~OD, data=Kelp)$resid
Kelp$LTD.g.W.OD<-lm(LTD~W+OD, data=Kelp)$resid
Kelp$BD.g.W.OD.LTD<-lm(BD~W+OD+LTD, data=Kelp)$resid W.g.OD= to capture the effect ofWthat is not shared withODLTD.g.W.OD= to capture the effect ofLTDthat is not shared withODorWBD.g.W.OD.LTD= to capture the effect ofBDnot shared withOD,W, orLTD
We can then fit a model that includes OD and all of these derived predictors:
seq.lm<-lm(Response~OD+W.g.OD+LTD.g.W.OD+BD.g.W.OD.LTD, data=Kelp)
summary(seq.lm)
Call:
lm(formula = Response ~ OD + W.g.OD + LTD.g.W.OD + BD.g.W.OD.LTD,
data = Kelp)
Residuals:
Min 1Q Median 3Q Max
-0.284911 -0.098861 -0.002388 0.099031 0.301931
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.747588 0.078192 35.139 < 2e-16 ***
OD 0.194243 0.028877 6.726 1.16e-07 ***
W.g.OD 0.008082 0.003953 2.045 0.0489 *
LTD.g.W.OD -0.055333 0.141350 -0.391 0.6980
BD.g.W.OD.LTD -0.004295 0.021137 -0.203 0.8402
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1431 on 33 degrees of freedom
Multiple R-squared: 0.6006, Adjusted R-squared: 0.5522
F-statistic: 12.41 on 4 and 33 DF, p-value: 2.893e-06To interpret the coefficients, we need to keep in mind how the predictors were constructed. For example, we would interpret the coefficient for OD as capturing unique contributions of OD as well as its shared contributions with W, LTD and BD. We would interpret the coefficient for W.g.OD as capturing the unique contributions of W as well as its shared contributions with LTD and BD that are not already accounted for by OD. The non-significant coefficients for LTD.g.W.OD and BD.g.W.OD.LTD could be interpreted as suggesting that the variables LTD and BD offer little to no new information about Response that is not already accounted for by OD and W.
Because of the way we created the variables, the variables are orthogonal - i.e., they are all uncorrelated. Thus, eliminating the lesser priority variables will not change the coefficients for the other variables.
seq.lm2<-lm(Response~OD+W.g.OD, data=Kelp)
summary(seq.lm2)$coef Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.747587774 0.076148241 36.082091 2.796476e-29
OD 0.194243475 0.028122800 6.906975 5.038589e-08
W.g.OD 0.008082141 0.003849614 2.099468 4.305538e-02It is important to recognize, however, that we are still unable to identify the unique contributions of the different variables, and that the coefficients we estimate depend on how we prioritize the predictors.
6.9 Principal components regression
Another way to create orthogonal predictors is to perform a principal component analysis (PCA) on the set of predictor variables under consideration. A PCA will create new orthogonal predictors from linear combinations5 of the original correlated variables:
where
Whereas the sequential regression approach from the last section created new orthogonal predictors through a prioritization and residual regression algorithm, a PCA creates new predictors such that
There are lots of functions in R that can be used to implement a PCA. Here, we use the princomp function in R (R Core Team 2021). PCAs formed using the covariance matrix of the predictor data will depend heavily on how variables are scaled (remember, we are finding linear combinations of the original predictor data that explain the majority of the variance in our predictors; if we do not scale our predictors first, or use the correlation matrix, then our principal components will be dominated by the variables that are largest in magnitude as these will have the largest variance). Thus, to avoid these issues, we add the argument cor=TRUE to indicate that we want to form PCAs using the correlation matrix (which effectively scales all predictors to have mean 0 and standard deviation of 1) rather than use the covariance matrix of our original predictor variables.
pcas<-princomp(~OD+BD+LTD+W, data=Kelp, cor=TRUE, scores=TRUE)
summary(pcas)Importance of components:
Comp.1 Comp.2 Comp.3 Comp.4
Standard deviation 1.6016768 0.8975073 0.60895215 0.50822166
Proportion of Variance 0.6413421 0.2013799 0.09270568 0.06457231
Cumulative Proportion 0.6413421 0.8427220 0.93542769 1.00000000We see that the first principal component accounts for 64% of the variation in the predictors, the second principal component accounts for 20% of the variation, and the other two together account for approximately 15% of the variation. We can also look at the loadings, which give the
pcas$loadings
Loadings:
Comp.1 Comp.2 Comp.3 Comp.4
OD 0.548 0.290 0.159 0.768
BD 0.545 0.179 0.581 -0.577
LTD -0.338 0.934
W 0.536 0.110 -0.795 -0.259
Comp.1 Comp.2 Comp.3 Comp.4
SS loadings 1.00 1.00 1.00 1.00
Proportion Var 0.25 0.25 0.25 0.25
Cumulative Var 0.25 0.50 0.75 1.00The new PCA variables are contained in pca$scores. We could also calculate these by “hand” if we first scaled and centered our original predictor data and then multiplied those values by the loadings as demonstrated below:
#scores by "hand" compared to scores returned by princomp
head(scale(as.matrix(Kelp[,2:5]))%*%pcas$loadings, n = 3) Comp.1 Comp.2 Comp.3 Comp.4
[1,] -0.1912783 -1.752736 0.66278941 -0.24694830
[2,] 0.6223409 -2.502387 -0.18091063 -0.46900655
[3,] -1.3326878 -0.919048 0.03361542 0.05590063 head(pcas$scores, n = 3) Comp.1 Comp.2 Comp.3 Comp.4
1 -0.1938459 -1.7762635 0.67168631 -0.25026319
2 0.6306949 -2.5359779 -0.18333907 -0.47530223
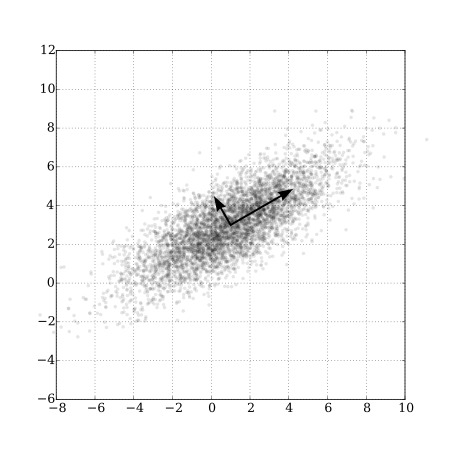
3 -1.3505770 -0.9313847 0.03406665 0.05665101As mentioned previously, it is common to plot the first two sets principal component scores along with the loadings, e.g., using the biplot function in R (Figure 6.8). From this plot, we can see that the first principal component is largely determined by OD, BD and W, whereas the second principal component is mostly determined by LTD. We can also arrive at this same conclusion by inspecting the loadings on each principal component. For example, if we inspect the loading for the 2nd principal component, we see that the largest loading is associated with LTD (0.934) with all other loadings being less than 0.3. Furthermore, we can see that the 3rd predictor contributes little to the 3rd and 4th principal components.
biplot(pcas)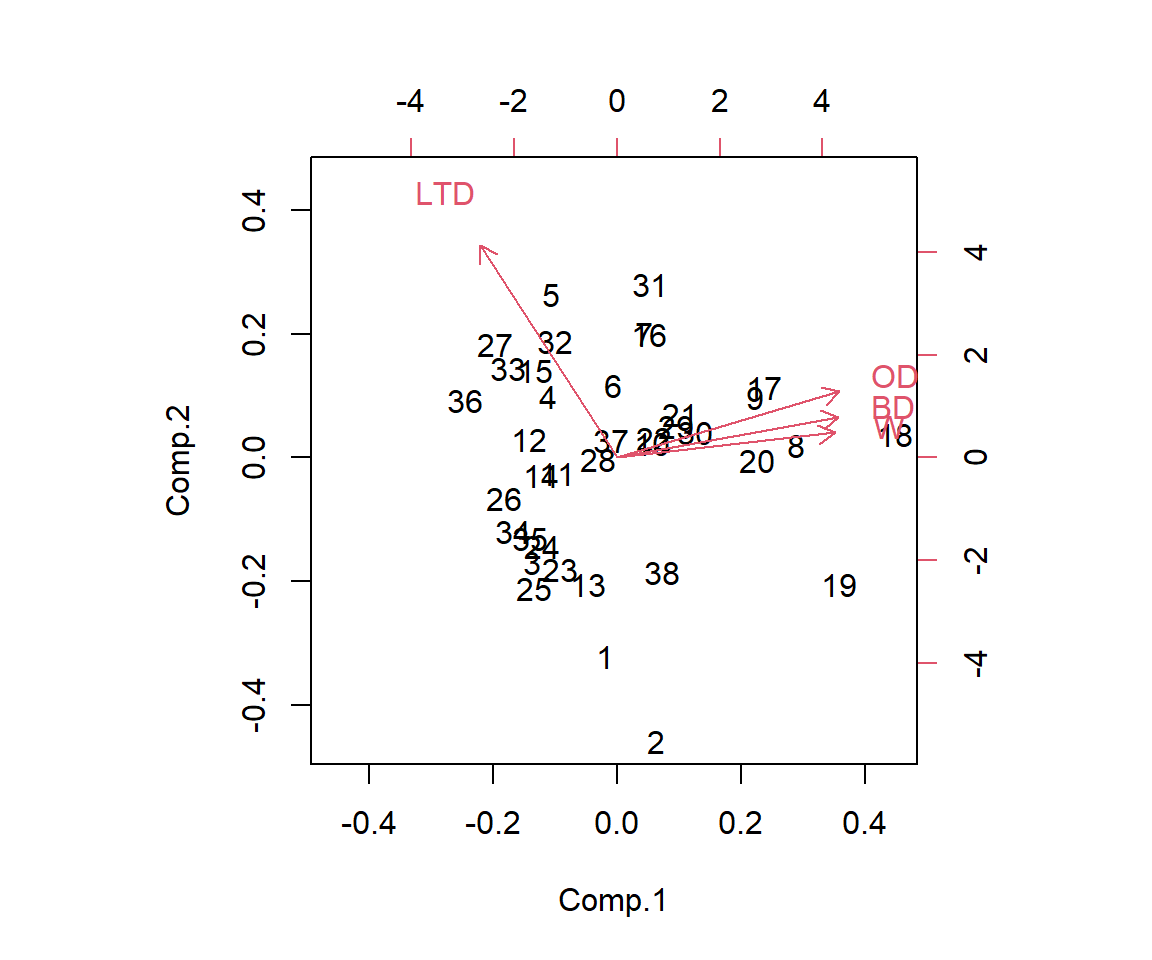
It is important to recognize that
Kelp<-cbind(Kelp, pcas$scores)
lm.pca<-lm(Response~ Comp.1 + Comp.2 + Comp.3 + Comp.4, data=Kelp)
summary(lm.pca)
Call:
lm(formula = Response ~ Comp.1 + Comp.2 + Comp.3 + Comp.4, data = Kelp)
Residuals:
Min 1Q Median 3Q Max
-0.284911 -0.098861 -0.002388 0.099031 0.301931
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.24984 0.02321 140.035 < 2e-16 ***
Comp.1 0.09677 0.01449 6.678 1.33e-07 ***
Comp.2 0.02931 0.02586 1.134 0.265
Comp.3 -0.03564 0.03811 -0.935 0.356
Comp.4 0.07722 0.04566 1.691 0.100
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1431 on 33 degrees of freedom
Multiple R-squared: 0.6006, Adjusted R-squared: 0.5522
F-statistic: 12.41 on 4 and 33 DF, p-value: 2.893e-06Since the
lm.pca2<-lm(Response ~ Comp.1, data = Kelp)
summary(lm.pca2)
Call:
lm(formula = Response ~ Comp.1, data = Kelp)
Residuals:
Min 1Q Median 3Q Max
-0.28269 -0.09537 0.00437 0.06927 0.35765
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.24984 0.02385 136.265 < 2e-16 ***
Comp.1 0.09677 0.01489 6.499 1.51e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.147 on 36 degrees of freedom
Multiple R-squared: 0.5398, Adjusted R-squared: 0.527
F-statistic: 42.23 on 1 and 36 DF, p-value: 1.507e-07Yet, if we used the coefficients and loadings associated with the remaining lm(Response ~ OD + W + LTD + BD). In the end, as a data-reduction technique (Harrell Jr 2015), we might choose to capture the combined effect of OD, W, LTD and BD using a single principal component (and 1 model degree of freedom). A downside of this approach is that this principal component can be difficult to interpret as it is a function of all of the original predictor variables. In addition, it may be useful to consider methods, such as partial least squares, that consider covariation between predictor and response variables when constructing orthogonal predictors (Scott and Crone 2021).
6.10 Other methods
There are several other multivariate regression techniques that one could consider when trying to eliminate collinear variables. In particular, one might consider combining similar variables (e.g., “weather variables”) using a PCA or variable clustering or scoring technique that creates an index out of multiple correlated variables (e.g., weather severity formed by adding a 1 whenever temperatures fall below, or snow depth falls above, pre-defined thresholds, DelGiudice et al. 2002; Dormann et al. 2013; Harrell Jr 2015). Scoring techniques are not that different from a multiple regression model, except rather than attempt to estimate optimal weights associated with each predictor (i.e., separate regression coefficients), variables are combined by assigning a +1, 0, -1 depending on whether the value of each predictor is expected to be indicative of larger or smaller values of the response variable, and then a single coefficient associated with the aggregated index is estimated.8 There are also several statistical methods (e.g., factor analysis, partial least squares, structural equation models, etc) that use latent variables to represent various constructs (e.g., personality, size, etc) that can be informed by or are the products of multiple correlated variables. We will not discuss these methods here, but note that they are popular in the social sciences. Some of these methods are touched on in Dormann et al. (2013).
6.11 References
Sometimes researchers will state that predictors were log-transformed to make them Normally distributed. Linear regression models do NOT assume that predictors are Normally distributed, so this reasoning is faulty. However, it is sometimes beneficial to consider log-transformations for predictor variables that have skewed distributions as a way to reduce the influence of individual outlying observations.↩︎
We will talk more about DAGs in Chapter 7, but for now, note that a DAG displays causal connections among a set of variables↩︎
It may seem strange to see “parameter estimator” here rather than “parameter estimates”. However, note that in statistics, bias is defined in terms of a difference between a fixed parameter and the average estimate across repeated samples (i.e., the mean of a sampling distribution). Thus, bias is associated with the method of generating estimates (i.e., the estimator), not the estimates themselves.↩︎
Graham (2003) suggests this ordering can be based on one’s instincts, intuition, or prior or current data, but how to use this information when deciding upon an order is not discussed in detail.↩︎
As a reminder, a linear combination is a weighted sum of the predictors.↩︎
For those readers that may have had a course in linear algebra, we can decompose
There are a multitude of multivariate regression methods, such as partial least squares, that focus instead on creating orthogonal variables that explain covariation between explanatory and response variables; see e.g., Scott and Crone (2021).↩︎
Jacob Cohen, a famous statistician, argued that we might be better off using these indices directly for inference rather than regression models when faced with a small number of observations and many correlated predictors; see Cohen (1992)↩︎