library(knitr) # for chunk options
library(kableExtra) # for tables
options(kableExtra.html.bsTable = T)
library(patchwork) # for multi-panel plots
library(ggplot2) # for plotting
library(dplyr) # for data wrangling
library(car) # for residual plots
library(R2jags) # for fitting models using JAGS
library(MCMCvis) # for visualizing MCMC output
library(modelsummary) # for summarizing models
library(MASS) # for generating multivariate random normal variables
library(ggeffects) # for summarizing fitted models
library(effects) # for summarizing fitted models
library(performance) # for residual plots
library(ggthemes) # for colorblind pallete 16 Logistic regression
Learning objectives
- Be able to formulate, fit, and interpret logistic regression models appropriate for binary data using R and JAGS
- Be able to compare models and evaluate model fit
- Be able to visualize models using effect plots
- Be able to describe statistical models and their assumptions using equations and text and match parameters in these equations to estimates in R output
16.1 R packages
We begin by loading a few packages upfront:
In addition, we will use data and functions from the following packages:
SightabilityModelfor the moose sightability datasospackage to search for functions to accomplish specific tasksResourceSelectionfor thehoslem.testfunctionLaplacesDemonfor student-t distributionglmxfor theBeetleMortalitydata setData4Ecologistsfor thewellsdata setlogistffor fitting a penalized likelihood version of a logistic regression model
16.2 Introduction to logistic regression
Often, we are interested in understanding or predicting binary response variables (i.e., variables that can take on only 1 of two values, Yes/No, Alive/Dead, Infected/Not-infected). Logistic regression is used to relate a binary response variable,
The quantity
- We are modeling
- We are modeling the logit of
- We are modeling the log odds of success (defined as
To determine
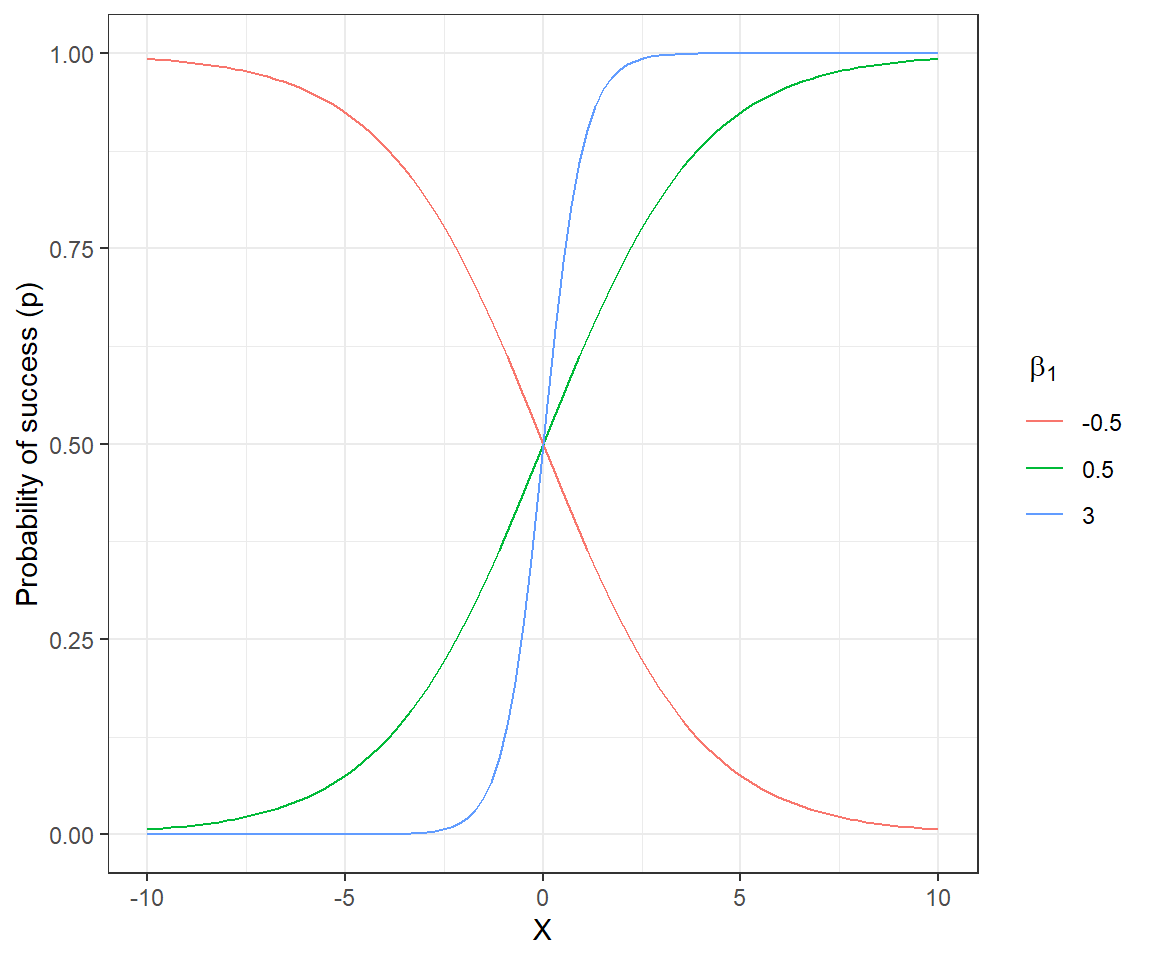
As the linear predictor,
We can gain further insights into the model by plotting
16.3 Parameter Interpretation: Application to moose detection data
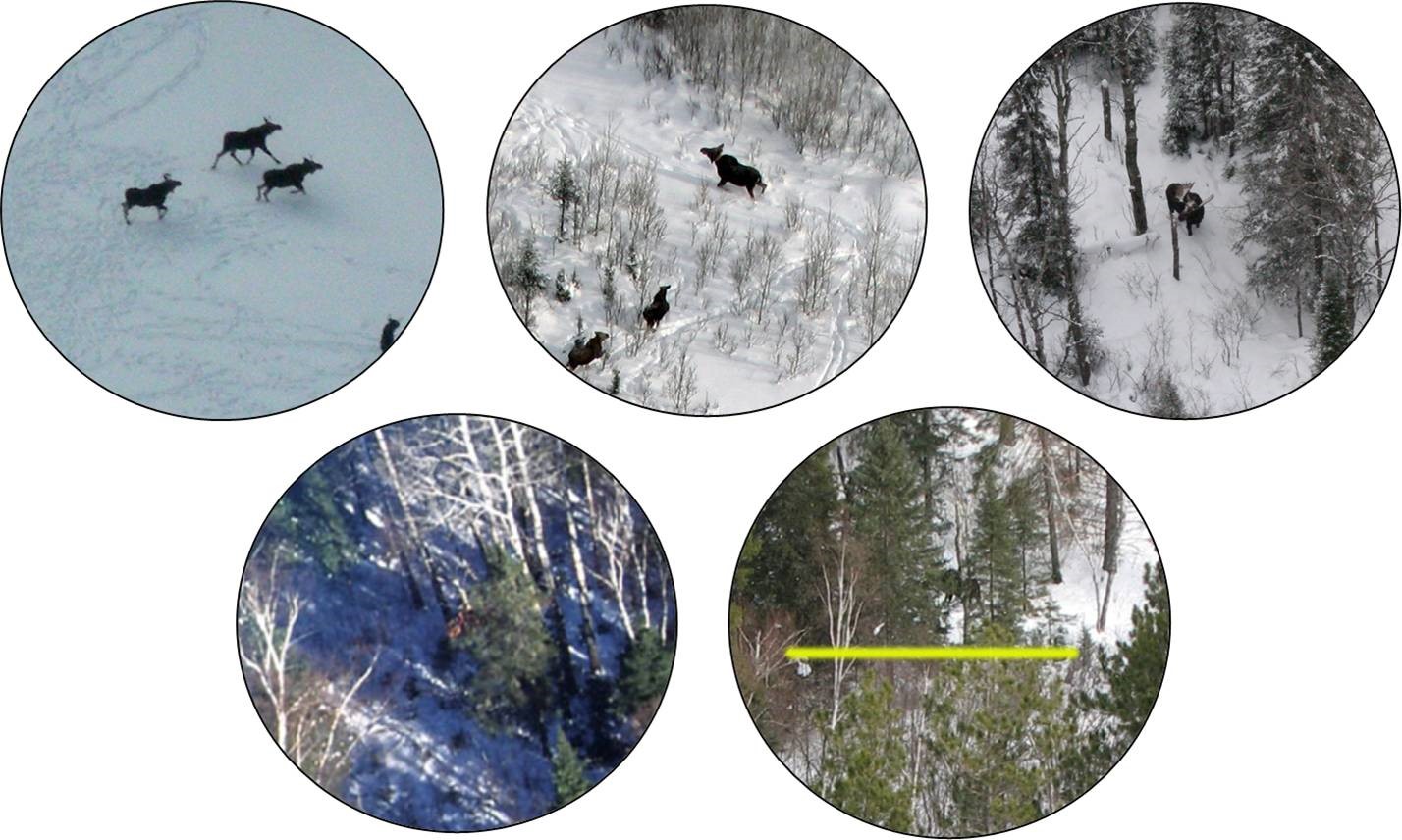
In Chapter 11, we considered data collected by the Minnesota Department of Natural Resoureces (MN DNR) to estimate the probability of detecting a moose when flying in a helicopter (Giudice, Fieberg, and Lenarz 2012). The probability of detecting a moose will depend on where the moose is located (e.g., the amount of cover that shields the moose from view, termed visual obstruction (or VOC); Figure 16.2). If we model the probability of detection as a function of VOC, we can then adjust counts of observed animals in future surveys that also record this information, providing a formal method for estimating moose abundance that accounts for imperfect detection (Fieberg 2012; Fieberg et al. 2013; ArchMiller et al. 2018). We might also want to consider whether detection probabilities vary by observer (a categorical variable) or whether detection probabilities varied among the four years of data collection.
Let’s read in a data set with the observations recording whether each moose was observed or not (observed) along with potential covariates for modeling the probability of detection. The data are contained in the SightabilityModel package (Fieberg 2012) and can be accessed using:
library(SightabilityModel)
data(exp.m)
str(exp.m)'data.frame': 124 obs. of 4 variables:
$ year : int 2005 2005 2005 2005 2005 2005 2005 2005 2005 2005 ...
$ observed: int 1 1 0 0 0 0 1 1 1 1 ...
$ voc : int 20 85 80 75 70 85 20 10 10 70 ...
$ grpsize : int 4 2 1 1 1 1 1 2 2 2 ...The data set in this package contains a small subset of the variables that could be used to model detection probabilities (see Giudice, Fieberg, and Lenarz 2012 for a discussion regarding how these were chosen). We will focus on the relationship between detection and visual obstruction measurements using the variables observed and voc, respectively. Note that the variable observed is binary:
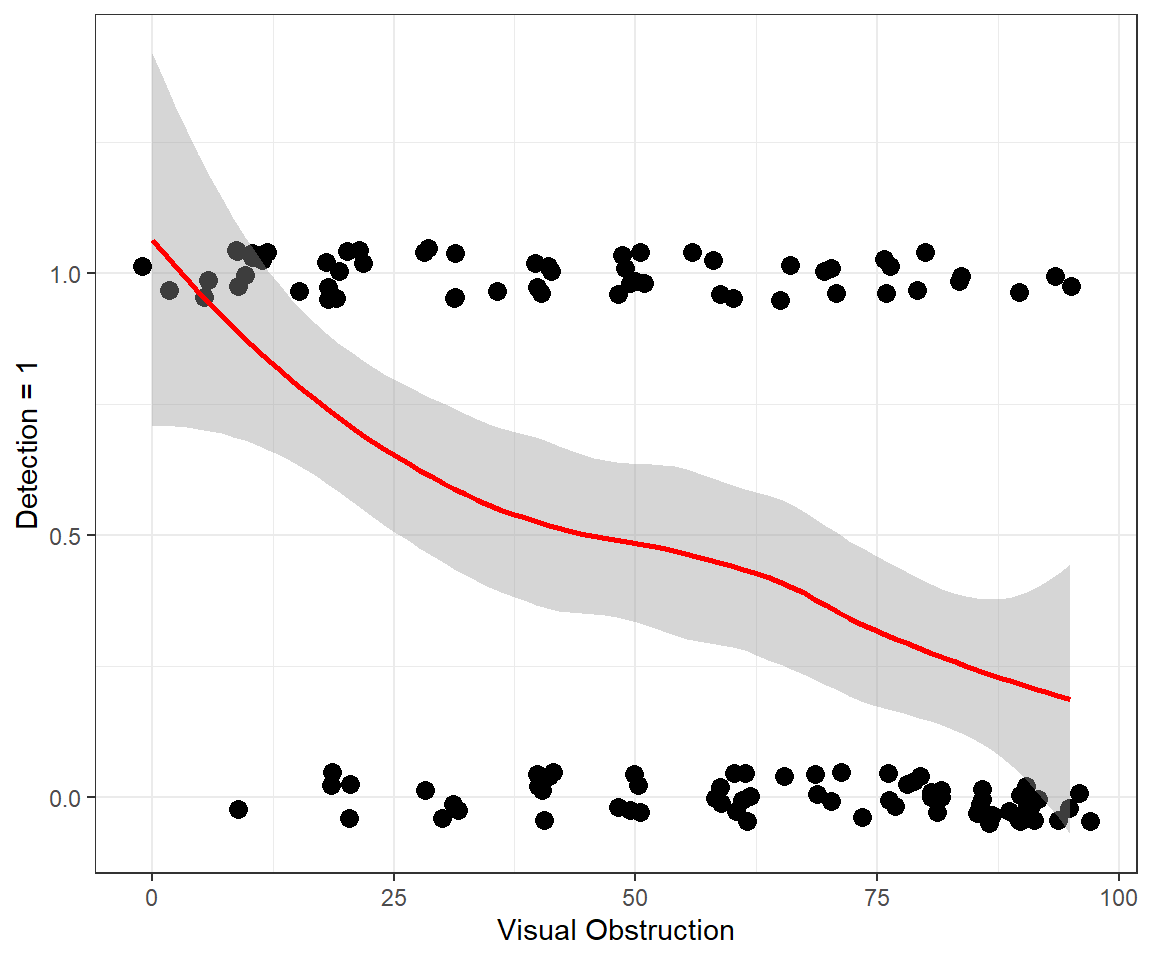
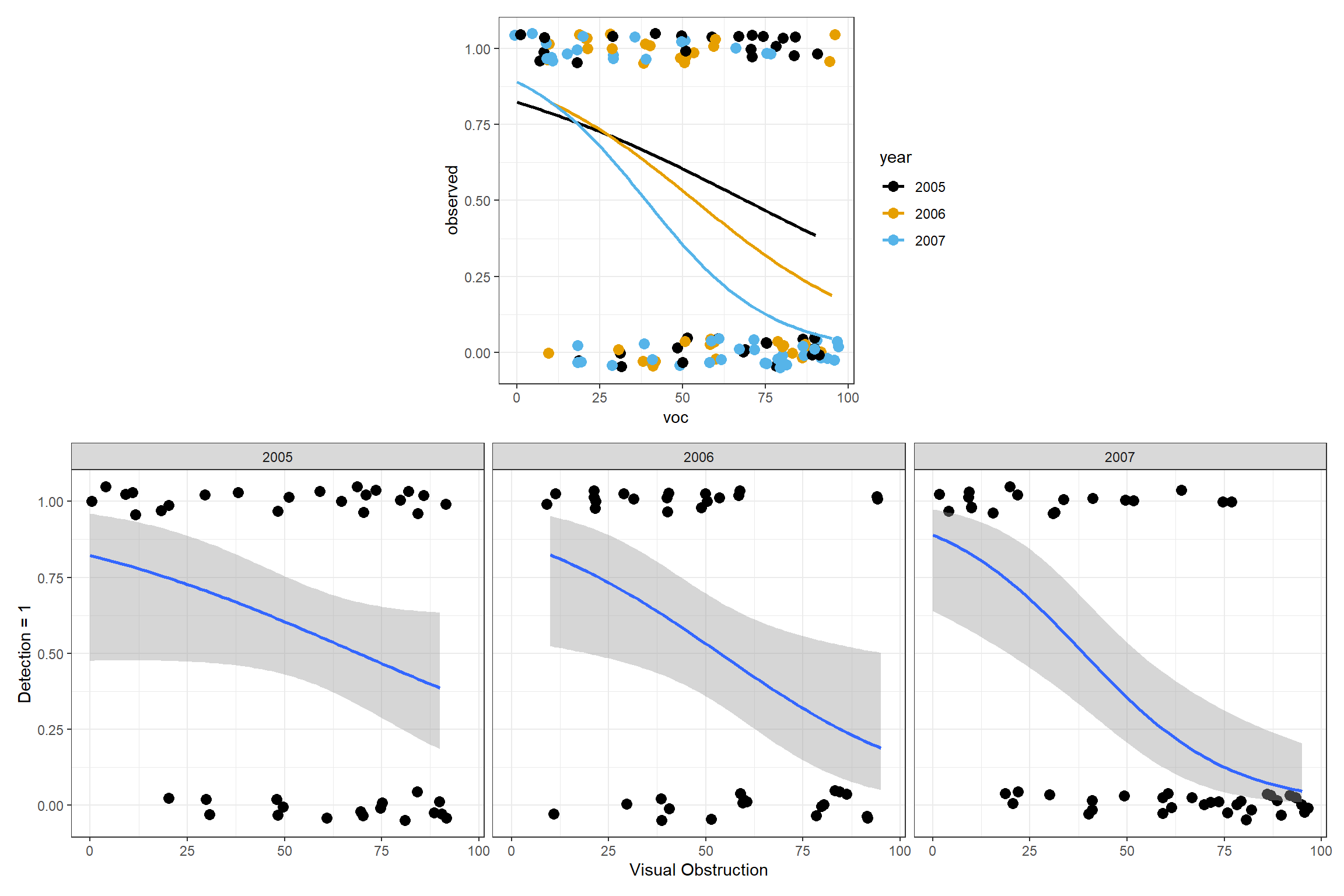
In Figure 16.3, we use a smoother to visualize overall trends in detection probabilities relative to voc. As expected, we see that detection probabilities decrease as the amount of screening cover increases. We could fit a linear regression model to the data. However, this approach would fail to capture several of the characteristics of the data:
- Detection probabilities must necessarily fall between 0 and 1.
- The observations do not have have constant variance (recall the variance of a Bernoulli random variable =
ggplot(exp.m, aes(voc,observed))+theme_bw()+
geom_point(position = position_jitter(w = 2, h = 0.05), size=3) +
geom_smooth(colour="red") + xlab("Visual Obstruction") +
ylab("Detection = 1")Letting
We can fit this model using glm with family =binomial().
mod1 <- glm(observed ~ voc, data = exp.m, family = binomial())By default, glm will use a logit link when specifying family=binomial(), but other links are possible. Similar to other regression models we’ve seen, we can use the summary function to view the estimated coefficients, their standard errors, and explore hypothesis tests for whether the coefficients are 0 (versus an alternative that they are not 0):
summary(mod1)
Call:
glm(formula = observed ~ voc, family = binomial(), data = exp.m)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.759933 0.460137 3.825 0.000131 ***
voc -0.034792 0.007753 -4.487 7.21e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 171.61 on 123 degrees of freedom
Residual deviance: 147.38 on 122 degrees of freedom
AIC: 151.38
Number of Fisher Scoring iterations: 4We see that the log-odds of detection decreases by -0.035 as we increase voc by 1 unit (i.e., for every increase of 1% visual obstruction). Alternatively, it is common to report effect sizes in terms of a change in odds. The odds of detection is given by:
Thus, if we increase voc by 1 unit, we increase the odds by glm model to calculate a 95% confidence interval for
beta1 <- coef(mod1)[2]
SEbeta1 <- sqrt(diag(vcov(mod1)))[2]
oddsr <- exp(beta1)
CI_beta <- beta1 + c(-1.96, 1.96)*SEbeta1
CI_odds <- exp(CI_beta)
estdata <- data.frame(beta1 = beta1, SEbeta1 = SEbeta1,
LCL_beta = CI_beta[1], UCL_beta = CI_beta[2], oddsratio = oddsr,
LCL_odds = CI_odds[1], UCL_odds = CI_odds[2])
colnames(estdata) <- c("$\\widehat{\\beta}_1$", "$SE(\\widehat{\\beta}_1)$",
"Lower 95\\% CL", "Upper 95\\% CL", "$\\exp(\\beta_1)$",
"Lower 95\\% CL", "Upper 95\\% CL")
kable(round(estdata,3), booktabs = TRUE, escape = FALSE)Alternatively, we could calculate profile-likelihood based confidence intervals by inverting the likelihood ratio test (Section 10.10). Recall, profile-likelihood intervals include all values, confint.
(ci.prof <- confint(mod1))Waiting for profiling to be done... 2.5 % 97.5 %
(Intercept) 0.89794673 2.71375978
voc -0.05082098 -0.02025314(exp(ci.prof)) 2.5 % 97.5 %
(Intercept) 2.4545581 15.0858887
voc 0.9504488 0.9799506The Normal-based and profile-likelihood confidence intervals are similar in this case. If confidence limits for
We can also use tab_model in the sjPlot library or the modelsummary function in the modelsummary package to calculate a table of effect sizes for our model (Table 16.3). By default, tab_model reports effect sizes in terms of odds, labeled Odds Ratios. We can can also calculate odds ratios by adding the argument exponentiate = TRUE when calling the modelsummary function (Table 16.3).
modelsummary(list("Logistic regression" = mod1),
exponentiate = TRUE, gof_omit = ".*", estimate = "{estimate} ({std.error})",
statistic=NULL,
coef_omit = "SD") To understand why voc with
What about the intercept? The intercept gives the log-odds of detection when voc = 0 (i.e., when moose are completely in the open). We can also use the plogis function in R to calculate the inverse-logit transform,
plogis(coef(mod1)[1]) (Intercept)
0.8532013 Thus, the model predicts there is an 85% chance of detecting a moose when it is completely in the open (i.e., when voc = 0).
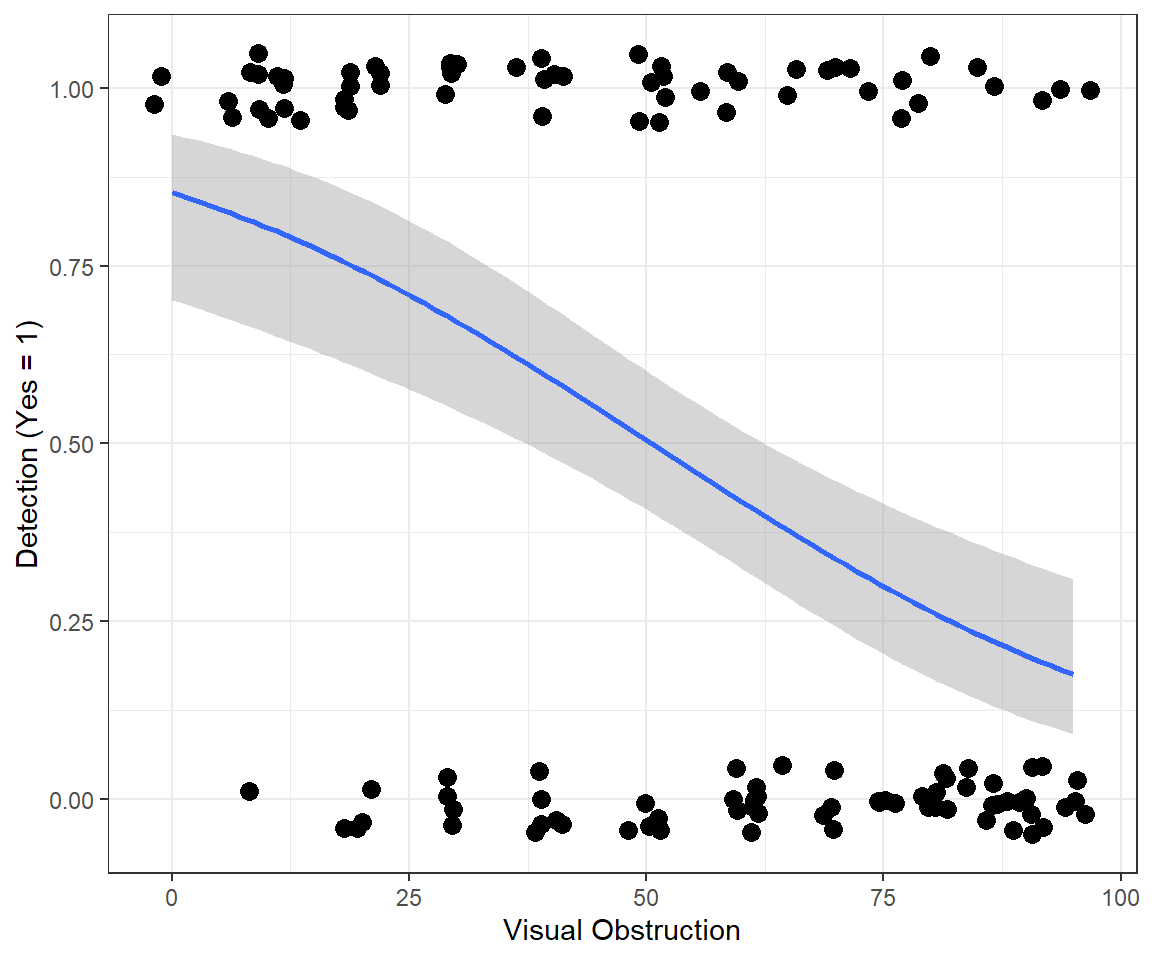
Lastly, we can visualize the model using ggplot, here with stat_smooth used to specify our regression model (Figure 16.4).
ggplot(exp.m, aes(voc,observed)) + theme_bw() +
geom_point(position = position_jitter(w = 2, h = 0.05), size=3) +
xlab("Visual Obstruction") +
stat_smooth(method="glm", method.args = list(family = "binomial") )+
ylab("Detection (Yes = 1)") 16.3.1 Model with continuous and categorical variables
Let’s now consider a second model, where we also include the year of the observation as a predictor. Rather than model a smooth trend in detection probabilities over time, we will use the as.factor function to create a categorical variable to represent the different years.
exp.m$year <- as.factor(exp.m$year)
mod2 <- glm(observed ~ voc + year, data = exp.m, family = binomial())
summary(mod2)
Call:
glm(formula = observed ~ voc + year, family = binomial(), data = exp.m)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.453203 0.622248 3.942 8.06e-05 ***
voc -0.037391 0.008199 -4.560 5.11e-06 ***
year2006 -0.453862 0.516567 -0.879 0.3796
year2007 -1.111884 0.508269 -2.188 0.0287 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 171.61 on 123 degrees of freedom
Residual deviance: 142.23 on 120 degrees of freedom
AIC: 150.23
Number of Fisher Scoring iterations: 4Again, R uses reference coding by default. This provides another opportunity to revisit design matrices from Chapter 3 as well as further test our ability to interpret the regression coefficients when we include multiple predictor variables.
Think-Pair-Share: How can we interpret the coefficients in our model with year and voc?
Since we did not include an interaction between voc and year, we have assumed the effect of voc is the same in all years. We can interpret the effect of voc in the same way as in Section 16.3, except we must now tack on the phrase while holding year constant. I.e., the log odds of detection changes by 0.037 and the odds of detection by 0.96 for every 1 unit increase in voc while holding year constant.
What about the three coefficients associated with the year variables that R created? These coefficients provide estimates of differences in the log odds of detection between each listed year and 2005 (the reference level), while holding voc constant. And, if we exponentiate these coefficients, we get ratios of odds (i.e., odds ratios) between each year and 2005 (Table 16.4), while holding voc constant.
Thus, for example, we can conclude that the odds of detection were 1/0.635 = 1.57 times higher in 2005 than 2006 if we hold voc constant.
16.3.2 Interaction model
What if we were to include an interaction between voc and year?
mod3 <- glm(observed ~ voc * year, data = exp.m, family = binomial())
summary(mod3)
Call:
glm(formula = observed ~ voc * year, family = binomial(), data = exp.m)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.53466 0.83338 1.841 0.0656 .
voc -0.02224 0.01287 -1.728 0.0839 .
year2006 0.35841 1.20786 0.297 0.7667
year2007 0.55335 1.13559 0.487 0.6261
voc:year2006 -0.01310 0.02002 -0.655 0.5127
voc:year2007 -0.03151 0.01983 -1.589 0.1121
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 171.61 on 123 degrees of freedom
Residual deviance: 139.60 on 118 degrees of freedom
AIC: 151.6
Number of Fisher Scoring iterations: 4Think-Pair-Share: How can we interpret the coefficients in the above model?
The year2006 and year2007 terms provide estimates of differences in intercepts between each listed year and the reference level (2005 in this case) and the voc:year2006 and voc:year2007 terms provide estimates of differences in slopes between each listed year and the reference level year. If this is not clear, it would help to review Chapter 3. You might also consider trying to write out the design matrix for a few observations in the data set that come from different years. You could then check your understanding using the model.matrix function. Try writing out the design matrix for the following observations:
exp.m[c(1,37,53,74), c("voc", "year")] voc year
37 20 2005
73 10 2007
89 60 2006
110 85 2007Check your answer by typing:
model.matrix(mod3)[c(1,37,53,74),]16.4 Evaluating assumptions and fit
16.4.1 Residual plots
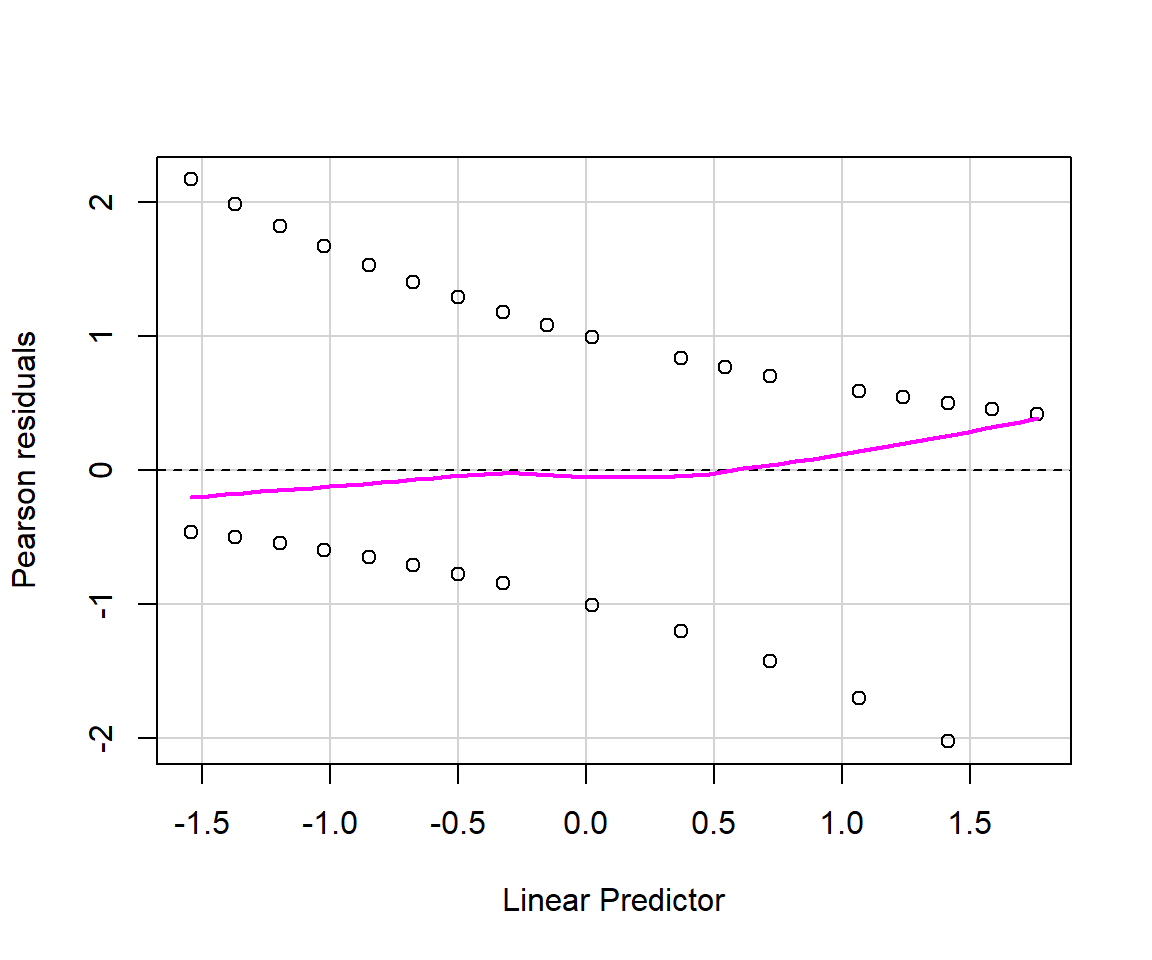
As discussed in Section 15.4.1, there are multiple types of residuals (standard, Pearson, deviance) that we might consider using to evaluate model fit. Below, we use the residualPlots function in the car library to create plots of Pearson residuals versus model predictors and versus fitted values (Figure 16.5).
residualPlot(mod1)The residuals for binary data always look weird since binned_residual function in the performance package will do this for us (Figure 16.6). It also provides error bounds and colors binned residuals that fall outside these bounds to help easily visualize whether the model provides a reasonable fit to the data.
binplot<-binned_residuals(mod1)
plot(binplot)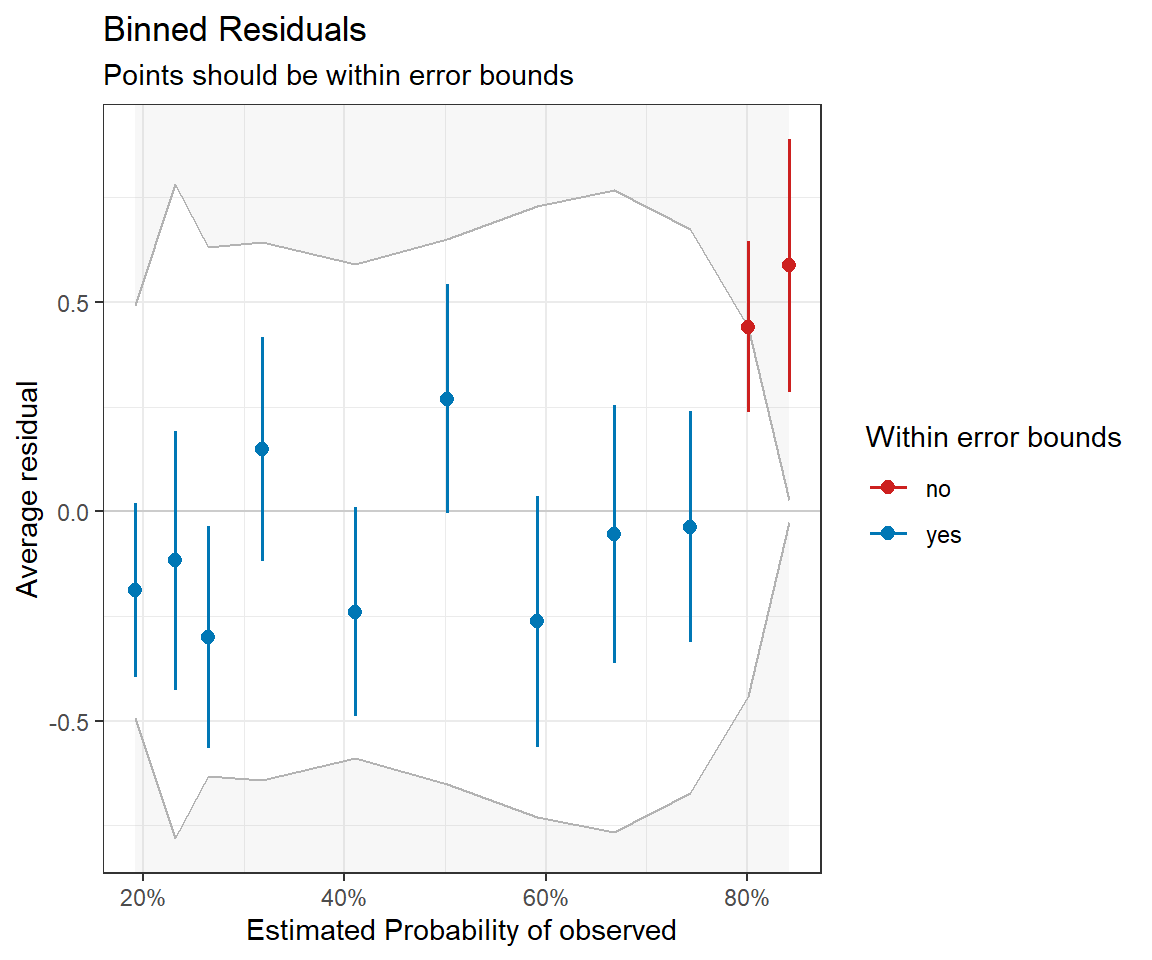
voc. The model looks OK, except that it is underpredicting those observations that have the highest probabilities of detection; these are the observations with the smallest values of voc.
16.4.2 Goodness-of-fit test
We can easily adapt the general approach for testing goodness-of-fit from Section 13.4 and Section 15.4.5 to logistic regression models. Again, we will consider the sum of Pearson residuals as our goodness-of-fit statistic:
For binary data analyzed using logistic regression, we have:
Here, we demonstrate the approach using the simpler model containing only voc.
nsims<-10000
gfit.sim<-gfit.obs<-matrix(NA, nsims, 1)
nobs<-nrow(exp.m)
beta.hat<-mvrnorm(nsims, coef(mod1), vcov(mod1))
xmat<-model.matrix(mod1)
for(i in 1:nsims){
# Form predictions using the randomly chosen betas
ps<-plogis(xmat%*%beta.hat[i,])
# Generate new y's
new.y<-rbinom(nobs, size=1, prob=ps)
# Calculate Pearson residuals
gfit.sim[i,]<-sum((new.y-ps)^2/(ps*(1-ps)))
gfit.obs[i,]<-sum((exp.m$observed-ps)^2/(ps*(1-ps)))
}
# GOF p-value
mean(gfit.sim > gfit.obs) [1] 0.466The p-value is much greater than 0.05, suggesting we do not have significant evidence to conclude that the logistic regression model with voc is inappropriate for our data.
16.4.3 Aside: alternative methods for goodness-of-fit testing
It is also common to see goodness-of-fit tests that ignore parameter uncertainty and that group observations to better meet the asymptotic
This approach is called the Hosmer-Lemeshow test (Hosmer Jr, Lemeshow, and Sturdivant 2013). There are many different implementations of this test in various R packages. To see a listing, we could use the findFn function from the sos package. If you run the code, below, you will see that there are over 40 packages that show up.
library(sos)
findFn("hosmer lemeshow")One such implementation is in the ResourceSelection package (Lele, Keim, and Solymos 2019).
library(ResourceSelection)
hoslem.test(exp.m$observed, fitted(mod1), g = 8)
Hosmer and Lemeshow goodness of fit (GOF) test
data: exp.m$observed, fitted(mod1)
X-squared = 3.2505, df = 6, p-value = 0.7768The performance_hosmer function in the performance package (Lüdecke et al. 2021) also implements this test.
performance_hosmer(mod1)# Hosmer-Lemeshow Goodness-of-Fit Test
Chi-squared: 6.674
df: 8
p-value: 0.572Summary: model seems to fit well.The p-values differ in the two implementations due to using a different number of groups (8 versus 10), which highlights a potential limitation of this test. Note, if we change g = 10 in the call to hoslem.test, we replicate the output from performance_hosmer.
16.5 Model comparisons
We can again use likelihood ratio tests to compare full and reduced models using either the drop1 function or the Anova function in the car package (Fox and Weisberg 2019). Let’s start with the model containing voc and year but not their interaction and consider whether we can simplify the model by dropping either voc or year.
drop1(mod2, test="Chisq")Single term deletions
Model:
observed ~ voc + year
Df Deviance AIC LRT Pr(>Chi)
<none> 142.23 150.23
voc 1 168.20 174.20 25.9720 3.464e-07 ***
year 2 147.38 151.38 5.1558 0.07593 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The top row of this table reports the AIC for the full model containing both voc and year. The second and third rows report AICs for reduced models in which either voc (second row) or year (third row) are dropped. The last two columns report the test-statistic and p-value associated with the likelihood ratio test comparing full and reduced models. The small p-value for the voc row suggests we should prefer the observed ~ voc + year model relative to the observed ~ year model. This again confirms that voc is an important predictor of the probability of detection. The results are less clear when comparing the observed ~ voc and observed ~ voc + year models, with a p-value of 0.07.
We can get the same set of tests using the Anova function in the car package:
Anova(mod2) Analysis of Deviance Table (Type II tests)
Response: observed
LR Chisq Df Pr(>Chisq)
voc 25.9720 1 3.464e-07 ***
year 5.1558 2 0.07593 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Lastly, we could compare nested or non-nested models using the AIC function. Here we compare:
- mod1 =
observed ~ voc - mod2 =
observed ~ voc + year - mod3 =
observed ~ voc * year
AIC(mod1, mod2, mod3) df AIC
mod1 2 151.3824
mod2 4 150.2266
mod3 6 151.6040The model with voc and year but not their interaction has the lowest AIC.
16.6 Effect plots: Visualizing generalized linear models
It is easy to use ggplot to visualize models containing a single continuous variable as in Figure 16.4. We can also use ggplot to easily visualize regression models that include 1 categorical and 1 continuous predictor, along with their interaction – using a single plot or a multi-panel plot (e.g., Figure 16.7).
p1<-ggplot(exp.m, aes(x=voc, y=observed, colour=year)) + theme_bw()+
geom_point(position = position_jitter(w = 2, h = 0.05), size=3) +
stat_smooth(method="glm", method.args = list(family = "binomial"),
se=FALSE) +
scale_colour_colorblind()
p2<-ggplot(exp.m, aes(voc,observed))+ theme_bw() +
geom_point(position = position_jitter(w = 2, h = 0.05), size=3) +
xlab("Visual Obstruction") +
stat_smooth(method="glm", method.args = list(family = "binomial"))+
ylab("Detection = 1") +facet_wrap("year")
(plot_spacer() + p1 + plot_spacer()) / p2 What if we fit more complicated models containing multiple predictor variables? We have a few options:
- We can create predicted values for different combinations of our explanatory variables, their standard errors, and then plot the results.
- We can use one or more R packages to automate this process. In Section 16.6.4, we will explore the
effects(Fox and Weisberg 2018) andggeffectspackages (Lüdecke 2018), which were also introduced in Section 3.14 when we discussed creating partial residual plots.
Let’s begin by creating our own plots, which will give us insights into how these latter packages work.
16.6.1 Predictions and confidence intervals
Let’s begin by considering our initial model containing only voc. We can summarize models in terms of: voc values (Figure 16.8). The easiest way to accomplish this is to use the predict function in R. Specifically, we can generate predictions for a range of values of voc using:
voc.p<-seq(1, 99, length = 100)
p.hat <- predict(mod1, newdata = data.frame(voc = voc.p), type = "response")plot(voc.p, p.hat, xlab = "VOC", ylab = "Pr(detect | voc)", type = "l")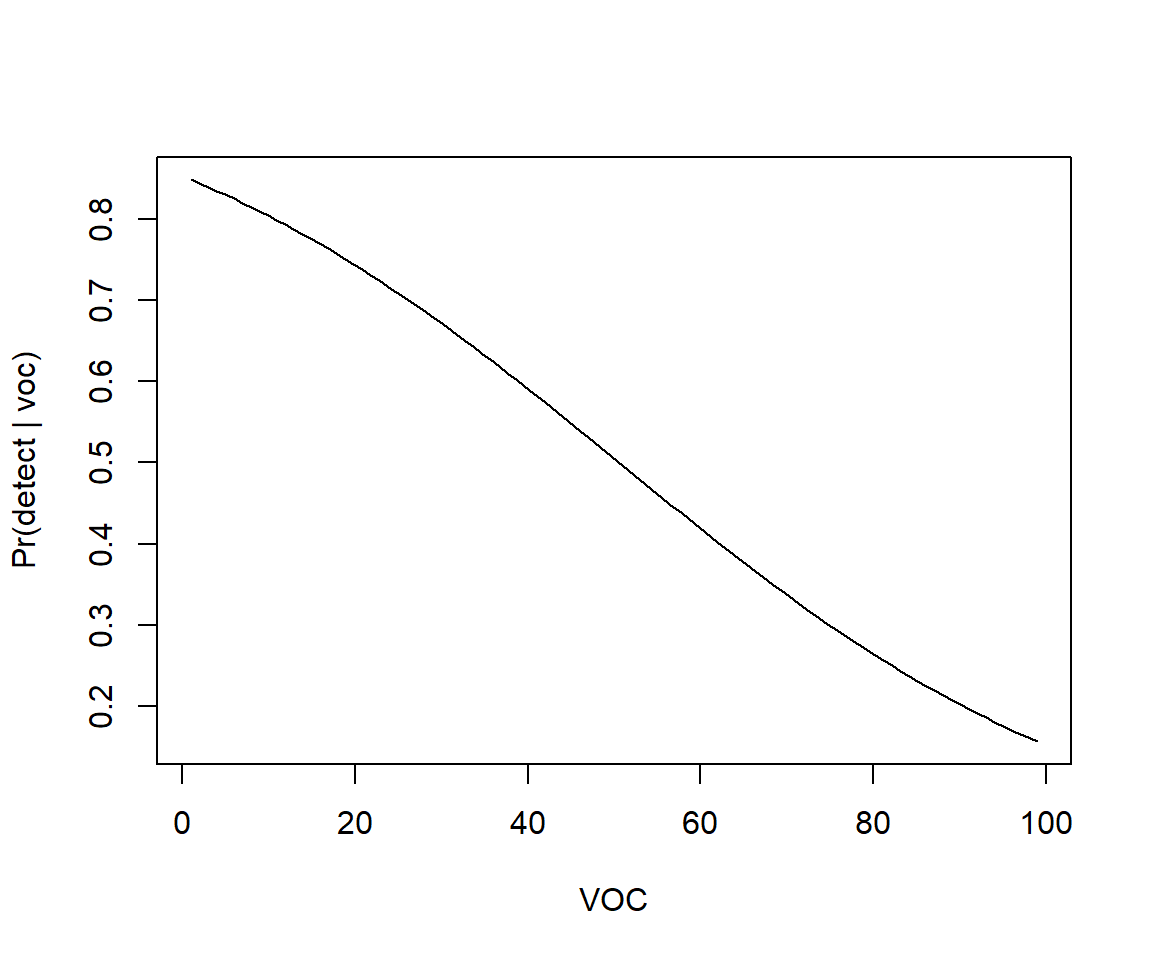
voc) only.
We could also ask the predict function to return standard errors associated with our predictions by adding the argument se.fit=TRUE. If we ask for SEs on the response scale, these standard errors will be calculated using the delta method (see Section 10.8.1). We could then use voc. Remember, that predict to generate estimates on the link (i.e., logit) scale, along with standard errors on this scale. Using this information, we can then form a confidence interval for plogis function to get a confidence interval for
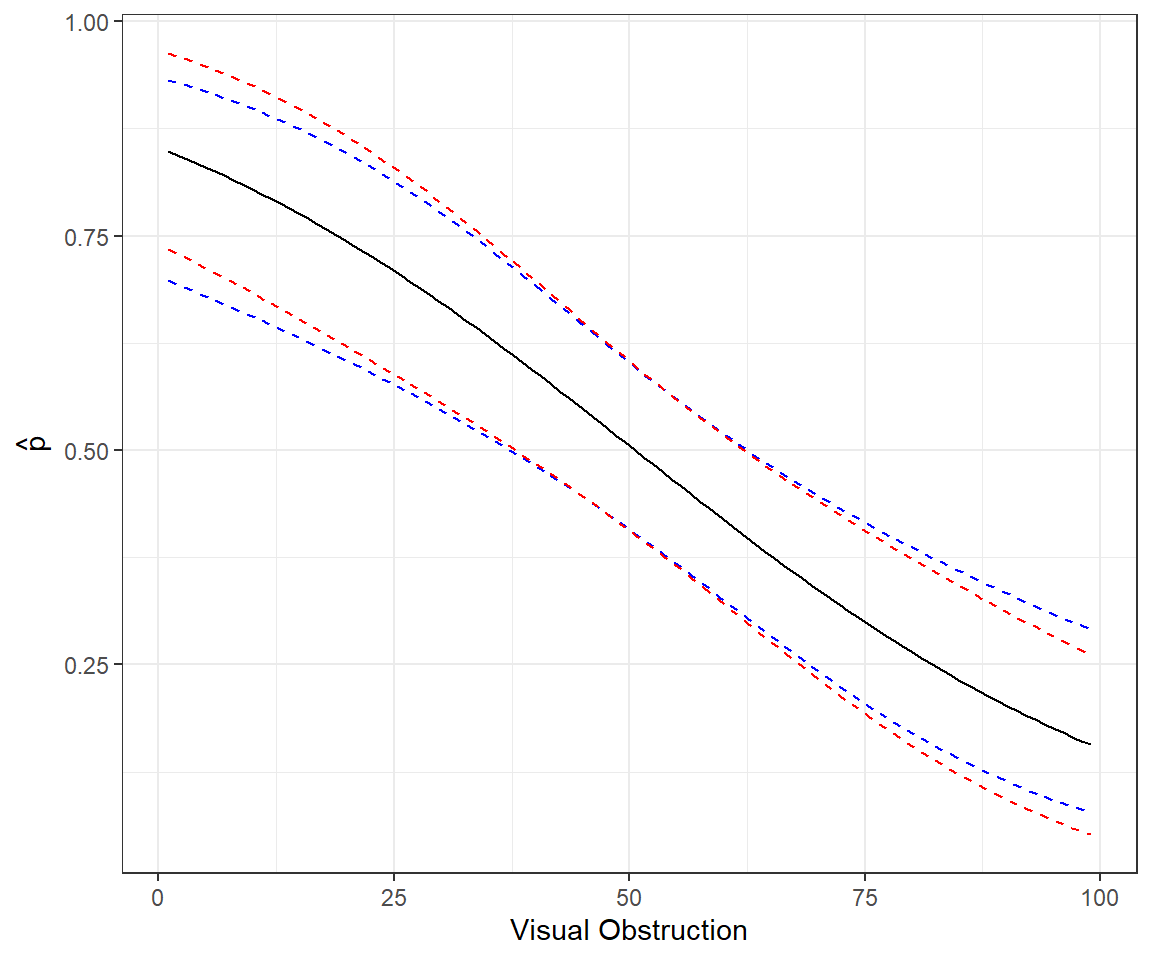
The sampling distribution for
Confidence intervals will be guaranteed to live on the (0,1) scale. Also, note that the intervals will not be symmetric.
The code and plot below compares these two approaches (Figure 16.9):
# Predictions response scale
newdat <- data.frame(voc = seq(1, 99, length = 100))
phat <- predict(mod1, type = "resp", se.fit = T,newdata = newdat)
lcl.r <- phat$fit + 1.96 * phat$se.fit
ucl.r <- phat$fit - 1.96 * phat$se.fit
# Predictions link scale
phat2<-predict(mod1, type = "link", se.fit = T, newdata = newdat)
lcl.l <- plogis(phat2$fit + 1.96 * phat2$se.fit)
ucl.l <- plogis(phat2$fit - 1.96 * phat2$se.fit)
pe.l <- plogis(phat2$fit)
# Combine and plot
newdat <- cbind(newdat, phat = phat$fit, lcl.r, ucl.r, phat2 = phat2$fit, lcl.l, ucl.l, pe.l)
ggplot(newdat, aes(voc, phat)) + geom_line() +
geom_line(aes(voc, lcl.l), lty = 2, col = "blue") +
geom_line(aes(voc, ucl.l), lty = 2, col = "blue") +
geom_line(aes(voc, lcl.r), lty = 2, col = "red") +
geom_line(aes(voc, ucl.r), lty = 2, col = "red") +
xlab("Visual Obstruction") + ylab(expression(hat(p)))16.6.2 Aside: Revisiting predictions using matrix algebra
How does R and the predict function work? Here, we briefly revisit the ideas from Section 5.6 and Section 3.12. Remember, glm estimates
Also, remember, for large samples, summary function) and to get confidence intervals.
Let voc measurements). Again, we can use model.matrix to see what this design matrix looks like.
Let
We can calculate
%*%in R to perform matrix multiplication)- Variance/covariance of
vcov(mod1)).
The code below demonstrates that we get the same results using matrix multiplication as when using the predict function.
newdata <- data.frame(observed = 1, voc = voc.p)
xmat <- model.matrix(mod1, data = newdata)
p.hat.check <- xmat %*% coef(mod1)
p.se.check <- sqrt(diag(xmat %*% vcov(mod1) %*% t(xmat)))
# Compare predictions
summary(predict(mod1, newdata = newdata, type = "link") - p.hat.check) V1
Min. :0
1st Qu.:0
Median :0
Mean :0
3rd Qu.:0
Max. :0 # Compare SEs
summary(predict(mod1, newdata = newdata, type = "link", se.fit = TRUE)$se.fit - p.se.check) Min. 1st Qu. Median Mean 3rd Qu. Max.
-5.551e-17 5.551e-17 5.551e-17 6.634e-17 1.110e-16 1.665e-16 16.6.3 Additive effects on logit scale
Creating and interpreting effect plots is a little trickier than it might seem due to modeling the mean on the logit scale. Let’s consider our second model:
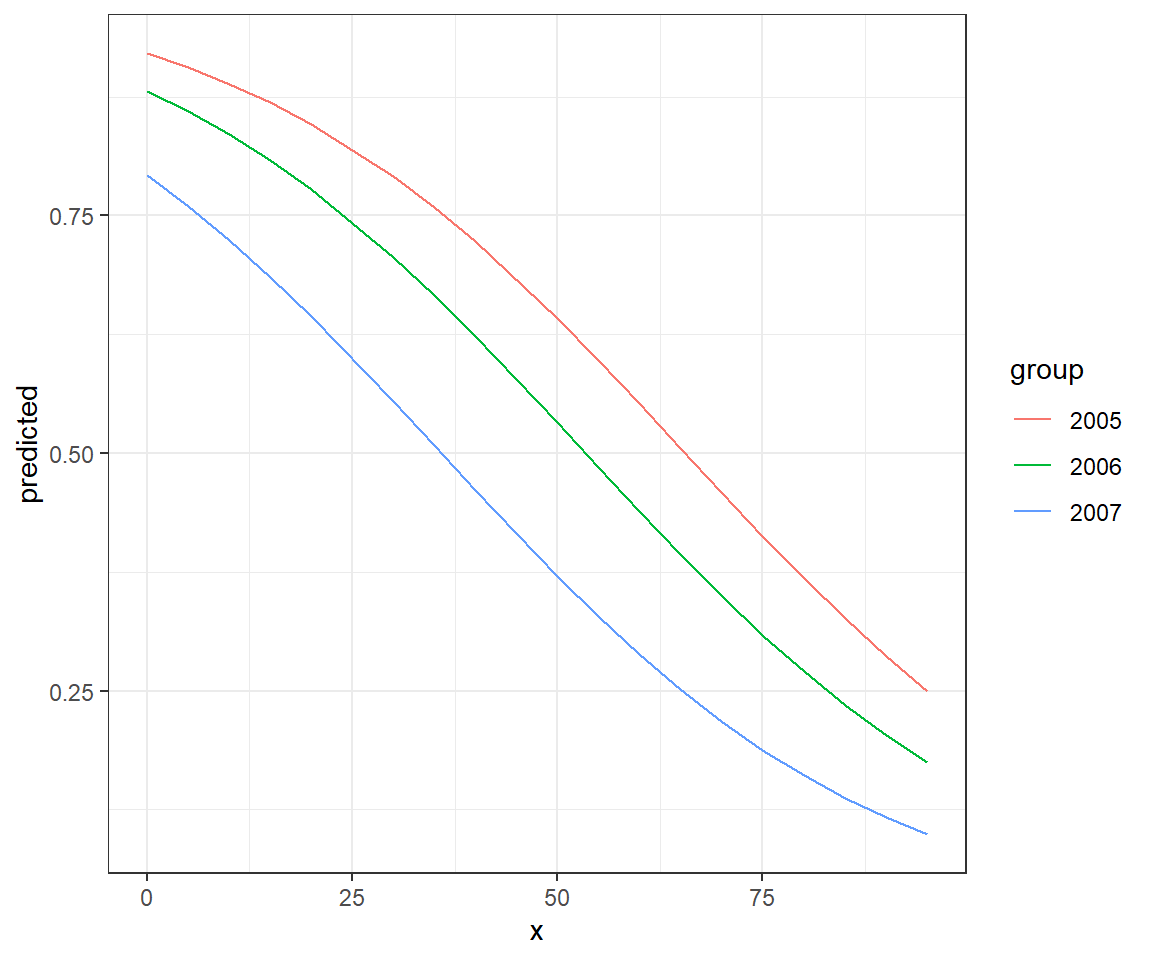
Here, the effects of year and voc are “additive” on the logit scale, and thus, differences in logit(voc. However, the effects of voc and year are multiplicative on the odds scale and even more complicated on the voc and the effect of voc on
To see this in action, let’s plot voc for the different years (Figure 16.10). Note that distances between the lines for the different years are constant when we look at
newdat<-expand.grid(voc=seq(0,100,5), year=unique(exp.m$year))
newdat$p.hats<-predict(mod2, newdat=newdat, type="response")
newdat$logit.p.hats<-predict(mod2, newdata=newdat, type="link")
plot.logitp<-ggplot(newdat, aes(voc, logit.p.hats, colour=year))+geom_line()+
ylab("logit(p)")
plot.p<-ggplot(newdat, aes(voc, p.hats, colour=year))+geom_line()+ylab("p")
plot.logitp + plot.p 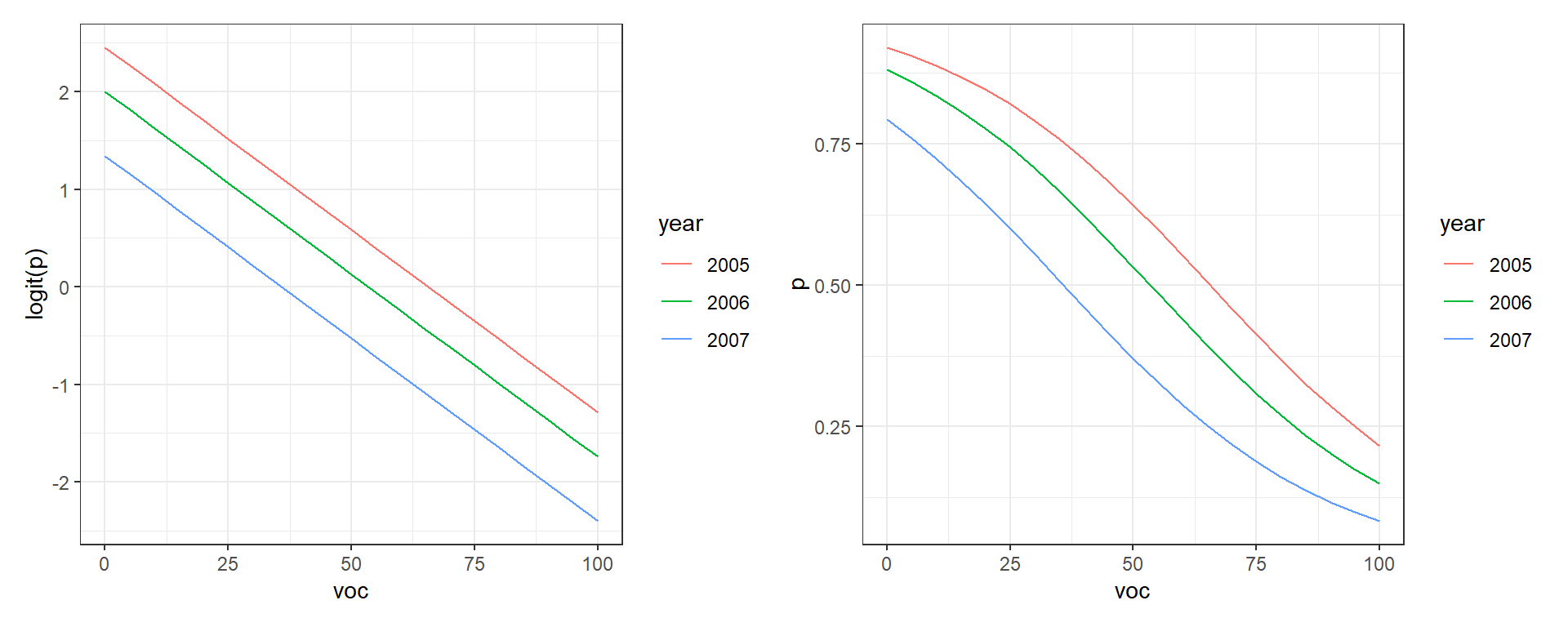
voc) and logit(16.6.4 Effect plots for complex models: Using the effects and ggpredict packages
Recall from Section 3.14.3, we can visualize the effect of predictors using effect plots formed by varying 1 predictor while holding all other predictors at a common value (e.g., the mean for continuous variables and the modal value for categorical variables). The expand.grid function, used in the creation of Figure 16.10 can be a big help here as it makes it easy to create a data set containing all combinations of multiple predictor variables. Still, creating effect plots from scratch can be a little tedious. The effects and ggeffects packages can makes this process a little easier.
16.6.4.1 Effects package
The effect function in the effects package:
- Fixes all continuous variables (other than the one of interest) at their mean values
- For categorical predictors, it averages predictions on the link scale, weighted by the proportion of observations in each category
By default, plots are constructed on the link scale, but we can add type="response" to create a plot on response scale. Examples are given in Figure 16.11 and Figure 16.12.
plot(effect("voc", mod2), type="response")
plot(effect("year", mod2), type="response")
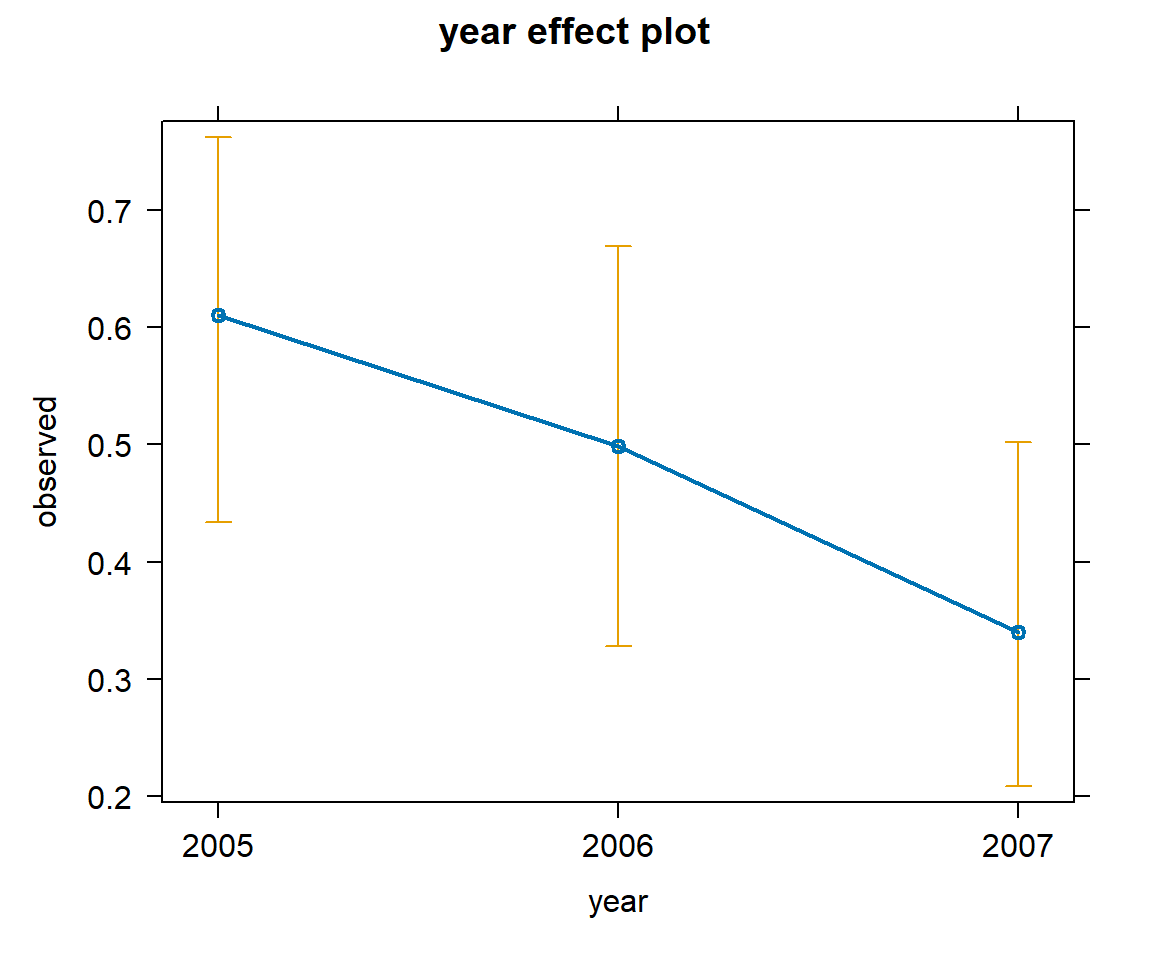
voc and with year using the model with voc and year.
We can add partial residuals using adding parital.residuals = TRUE to the effect function (Figure 16.12). For binary data, it is difficult to interpret the raw residuals, but this option also overlays a smooth line capturing any trend.
plot(effect("voc", mod2, partial.residuals = TRUE), type="response")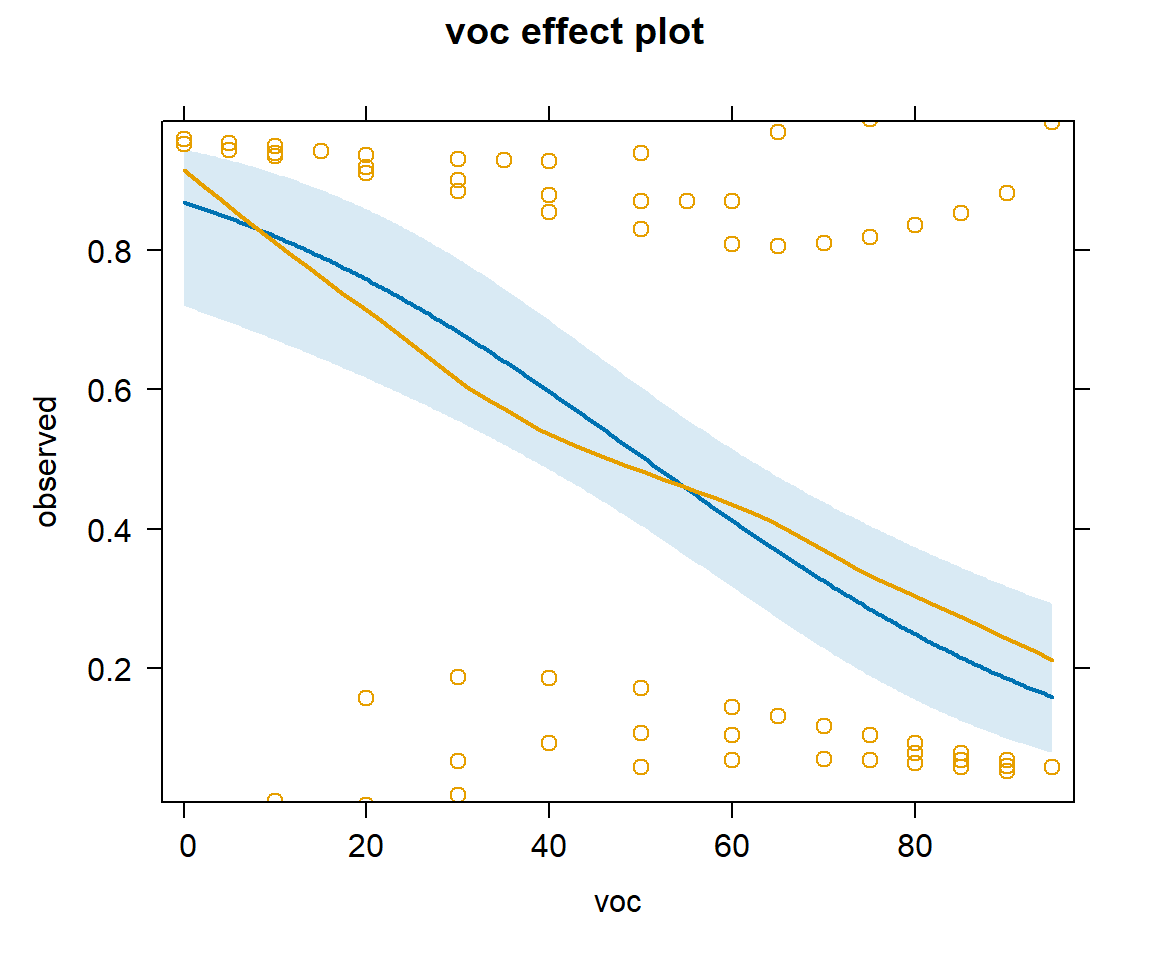
voc using the model with voc and year.
We can also use effect and the summary functions to return numerical values (estimates and confidence intervals):
summary(effect("year", mod2))
year effect
year
2005 2006 2007
0.6105102 0.4988990 0.3401946
Lower 95 Percent Confidence Limits
year
2005 2006 2007
0.4338786 0.3283351 0.2086299
Upper 95 Percent Confidence Limits
year
2005 2006 2007
0.7622325 0.6697194 0.5020875 Alternatively, we can use the ggeffect function in the ggeffects package to format the output as a list with an associated print function.
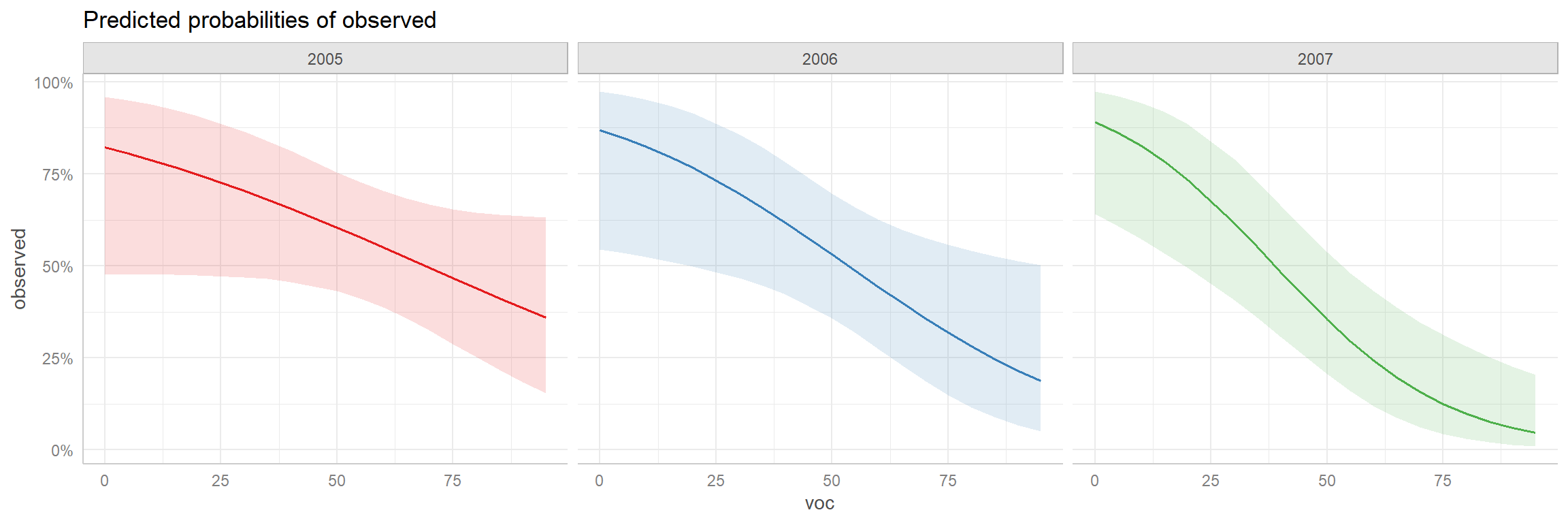
ggeffect(mod2)$voc
# Predicted probabilities of observed
voc | Predicted | 95% CI
----------------------------
0 | 0.87 | 0.72, 0.94
10 | 0.82 | 0.67, 0.91
20 | 0.76 | 0.62, 0.86
35 | 0.64 | 0.52, 0.74
55 | 0.46 | 0.36, 0.56
65 | 0.37 | 0.27, 0.47
75 | 0.29 | 0.19, 0.41
95 | 0.16 | 0.08, 0.29
Not all rows are shown in the output. Use `print(..., n = Inf)` to show
all rows.
$year
# Predicted probabilities of observed
year | Predicted | 95% CI
-----------------------------
2005 | 0.61 | 0.43, 0.76
2006 | 0.50 | 0.33, 0.67
2007 | 0.34 | 0.21, 0.50
attr(,"class")
[1] "ggalleffects" "list"
attr(,"model.name")
[1] "mod2"There is also a plot function that we can use with the ggeffect function to visualize these effects (Figure 16.13).
p1 <- plot(ggeffect(mod2, "year"))
p2 <- plot(ggeffect(mod2, "voc"))
p1 + p2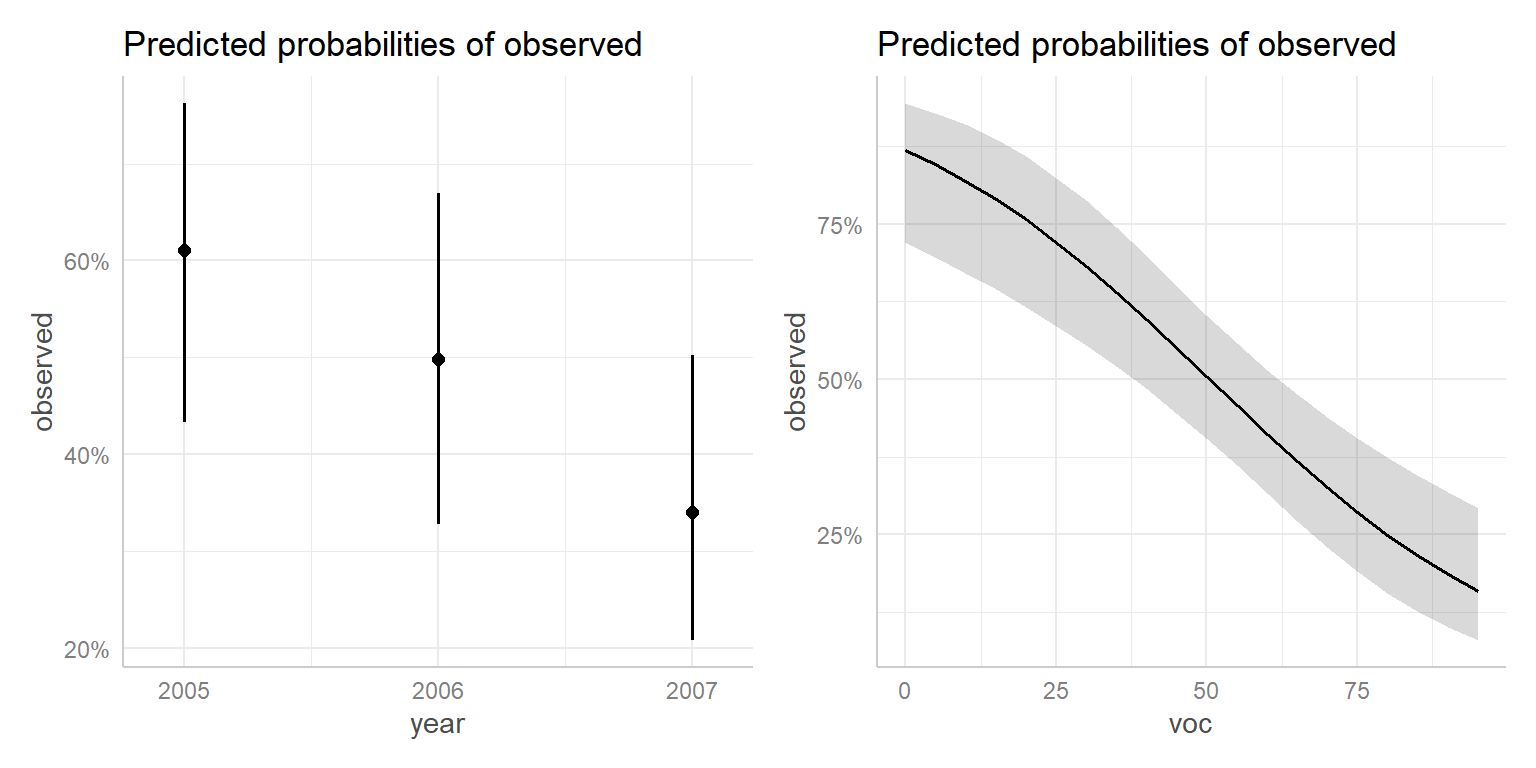
voc and year created using the ggeffects package.
And again, we could add partial residuals and a smooth line to our plot (Figure 16.14).
plot(ggeffect(mod2, "voc"), show_residuals = TRUE, show_residuals_line = TRUE)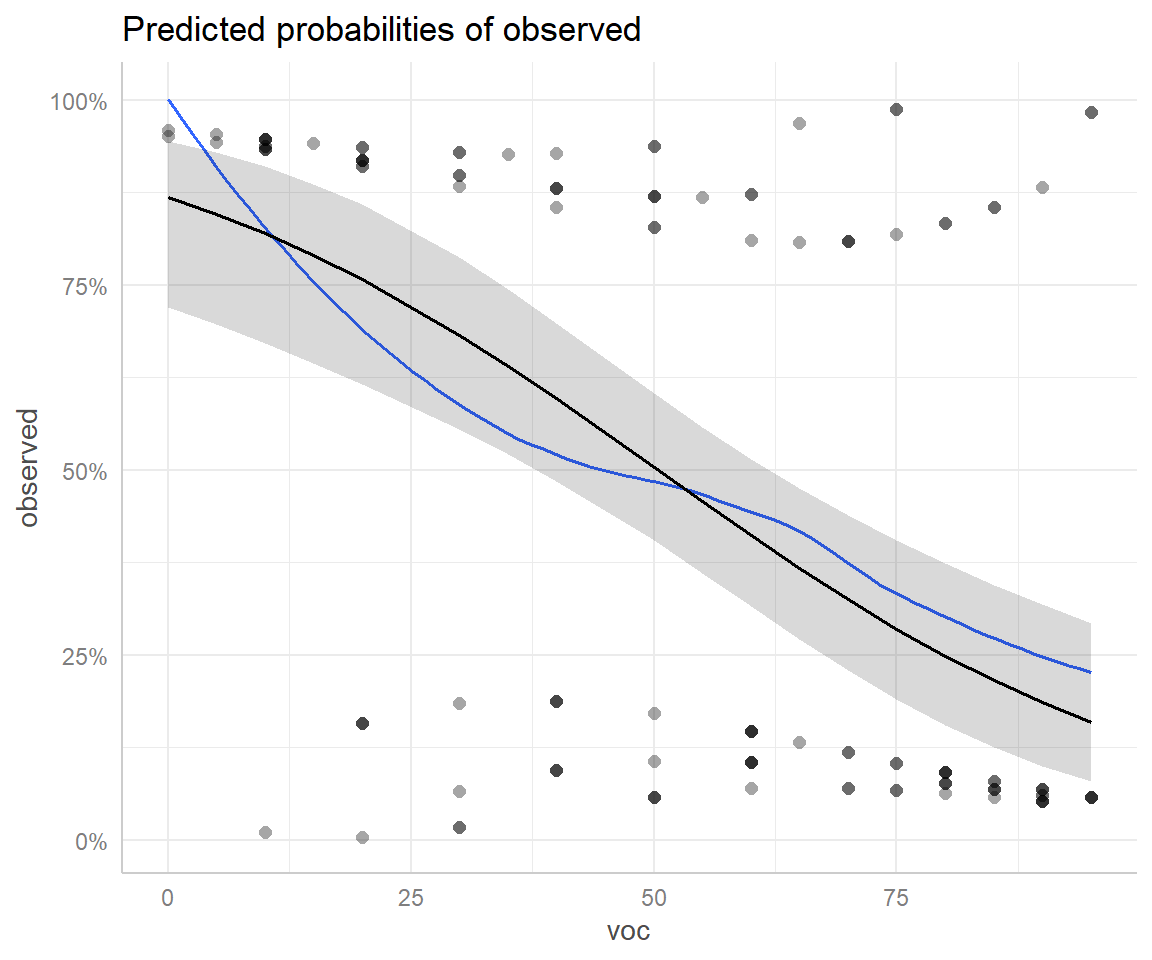
ggeffects package with partial residuals overlayed.
Lastly, we can specify specific terms for which we want estimates. In this case, ggeffects will return a data.frame from which we can create our own plot using ggplot (Figure 16.15).
16.6.5 Understanding effect estimates from the effects and ggeffects packages
The goal of this section is to illuminate how the estimates and confidence intervals on the response scale are formed when using the effects package or the ggeffects function in the ggeffects package. Let’s begin by considering the effects for year. These are formed by setting voc to its mean value, and estimating:
predict function:
newdata<-data.frame(voc = rep(mean(exp.m$voc), 3),
year = c("2005", "2006", "2007"))
predict(mod2, newdata = newdata, type = "resp") 1 2 3
0.6105102 0.4988990 0.3401946 effect("year", mod2, type = "response")
year effect
year
2005 2006 2007
0.6105102 0.4988990 0.3401946 Now, how does the effects package estimate the effect of voc? There is no “mean or average” year since year is a categorical variable. Instead, it calculates a weighted mean of the predictions for each year on the link scale, with weights given by the proportion of observations in each category. It then back-transforms this weighted mean.
effect("voc", mod2)
voc effect
voc
0 20 50 70 100
0.8684553 0.7575962 0.5044524 0.3251918 0.1356677 # Proportion of observations in each year (will be used to form weights)
weights <- data.frame(table(exp.m$year) / nrow(exp.m))
names(weights) = c("year", "weight")
weights year weight
1 2005 0.3145161
2 2006 0.2983871
3 2007 0.3870968# New data for predictions
newdata <- data.frame(expand.grid(year = c("2005", "2006", "2007"),
voc = seq(0, 100, 20)))
newdata <- left_join(newdata, weights)
# Predictions on link scale
newdata$logit.phat <- predict(mod2, newdata = newdata, type = "link")
# Weight predictions and backtransform
newdata %>% group_by(voc) %>% dplyr::summarize(plogis(sum(logit.phat * weight)))# A tibble: 6 × 2
voc `plogis(sum(logit.phat * weight))`
<dbl> <dbl>
1 0 0.868
2 20 0.758
3 40 0.597
4 60 0.412
5 80 0.249
6 100 0.13616.6.6 Predictions using the ggpredict function
Whereas the effects function averages predictions across the different levels of a categorical predictor, producing what are sometimes referred to as marginal effects, the ggpredict function will provide adjusted predictions where all variables except a focal variable remain fixed at their mean, modal, or user-specified values. We first demonstrate this approach for our additive model with voc and year.
ggpredict(mod2)$voc
# Predicted probabilities of observed
voc | Predicted | 95% CI
----------------------------
0 | 0.92 | 0.77, 0.98
10 | 0.89 | 0.73, 0.96
20 | 0.85 | 0.67, 0.94
35 | 0.76 | 0.58, 0.88
55 | 0.60 | 0.42, 0.75
65 | 0.51 | 0.34, 0.67
75 | 0.41 | 0.25, 0.59
95 | 0.25 | 0.12, 0.45
Adjusted for:
* year = 2005
Not all rows are shown in the output. Use `print(..., n = Inf)` to show
all rows.
$year
# Predicted probabilities of observed
year | Predicted | 95% CI
-----------------------------
2005 | 0.55 | 0.38, 0.71
2006 | 0.44 | 0.28, 0.62
2007 | 0.29 | 0.17, 0.45
Adjusted for:
* voc = 60.00
attr(,"class")
[1] "ggalleffects" "list"
attr(,"model.name")
[1] "mod2"As with the ggeffects function, if we specify terms from the model, the function will return a data.frame that can be easily plotted. This approach can be particularly useful when there are interactions between variables as in our mod3 (Figure 16.16).
16.7 Logistic regression: Bayesian implementations
Again, an advantage of implementing models in JAGS is that we will be forced to be very clear about the model we are fitting. Specifically, we will have to write down the likelihood with the expression relating glm where users may not even know that the mean is being modeled on a transformed scale.
In this section, we will fit the logistic regression model containing only voc, leaving the model with year and voc as an in-class exercise. Before we look at JAGS code for fitting this model, it is helpful to give some thought to priors for our regression parameters. It turns out that specifying priors can be a bit tricky. We can specify priors that are vague (meaning they take on a wide range of values, all equally likely) on the logit scale, but this may then imply something very specific when viewed on the
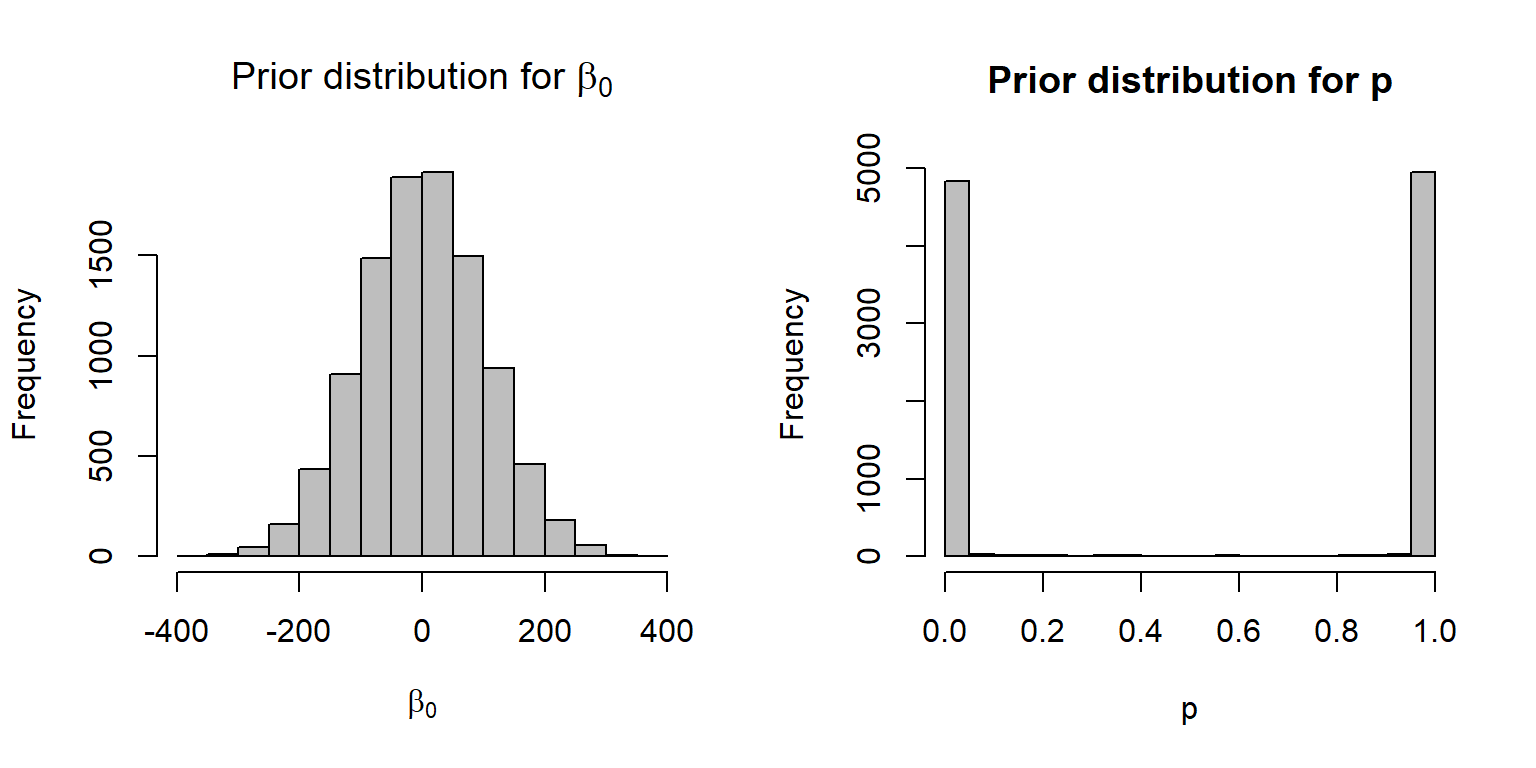
It turns out (see Figure 16.17) that this prior is not at all vague when viewed on the
par(mfrow=c(1,2))
beta0<-rnorm(10000,0,100)
p<-plogis(beta0)
hist(beta0, xlab=expression(beta[0]),
main=expression(paste("Prior distribution for ", beta[0])),
col="gray")
hist(p, xlab="p", main="Prior distribution for p", col="gray")If the goal is to achieve a near uniform distribution for
par(mfrow=c(1,2))
beta0<-rnorm(10000, 0, sqrt(3))
p<-plogis(beta0)
hist(beta0, xlab=expression(beta[0]),
main=expression(paste("Prior distribution for ", beta[0])),
col="gray")
hist(p, xlab="p", main="Prior distribution for p", col="gray")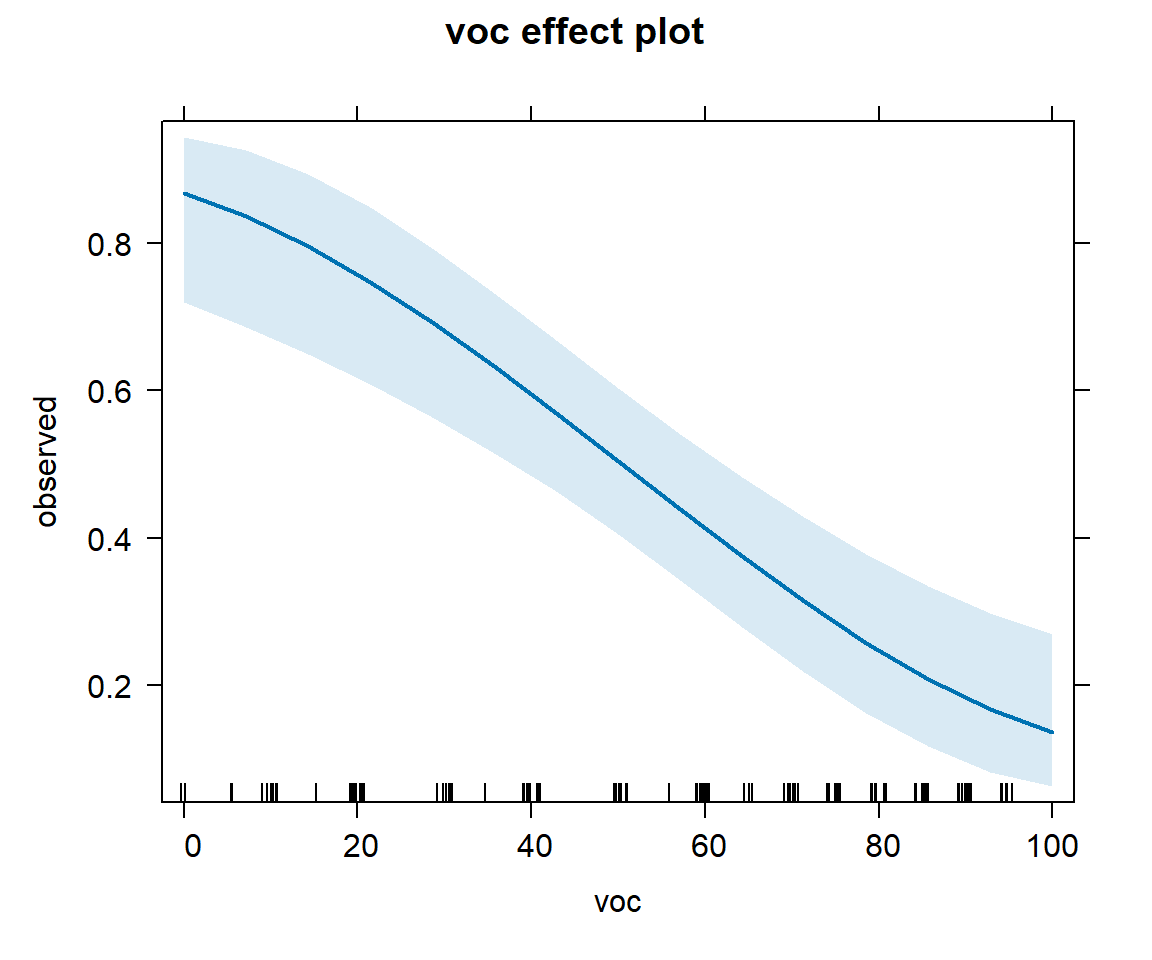
Establishing recommendations for default priors is a high priority and active research area within the Bayesian community (see discussion here: https://github.com/stan-dev/stan/wiki/Prior-Choice-Recommendations). For logistic regression models, Gelman et al. (2008) recommended:
- Scaling continuous predictors so they have mean 0 and sd = 0.5.
- Using a Cauchy prior, with precision parameter =
dt(0, pow(2.5,-2), 1).
Both
library(LaplacesDemon)
curve(dnorm(x, mean=0, sd=2.5), from=-8, to=8, ylab="Density")
curve(dstp(x, mu=0, tau=1/2.5^2, n=1), from=-8, to=8, add=TRUE, lty=2)
legend(2, 0.15, c("Normal", "t"), lty=c(1,2), bty="n")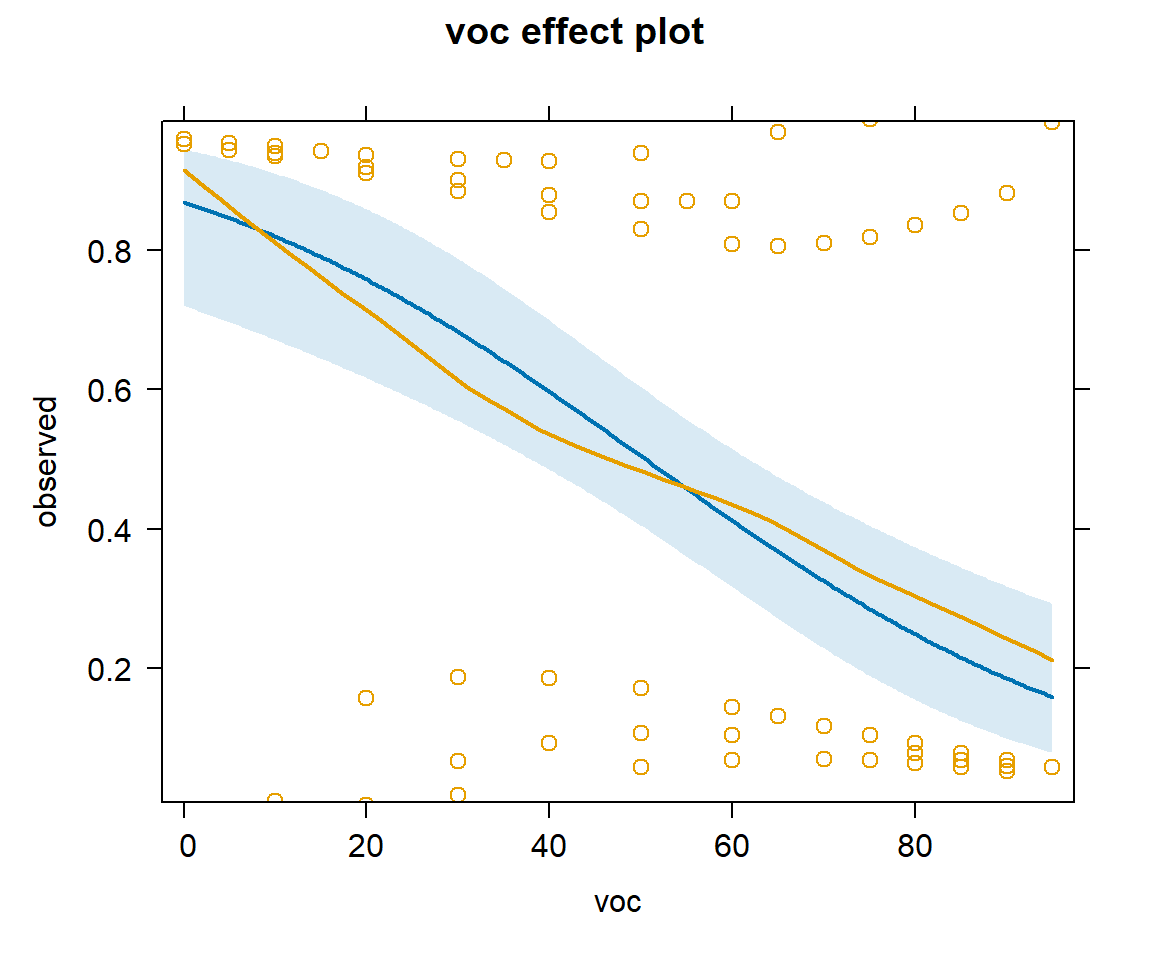
In general, we may seek a non-informative prior, which means that we want our answers to be determined primarily by the data and likelihood and not the prior. On the other hand, an advantage of a Bayesian approach is that we can potentially take advantage of previous knowledge and data when this information exists and we want it to influence our results. I.e., there may be times when using an informative prior can be beneficial. Furthermore, many would argue for weakly informative priors that “regularize” or shrink parameters towards 0 (e.g., McElreath 2020). This approach is often used to improve predictive performance, particularly in cases where one is faced with having too many explanatory variables relative to one’s sample size (Section 8.7).
16.7.1 Fitting the Bayesian logistic regression model to moose sightability data
Below, we use Gelman’s suggested prior when analyzing the moose data in JAGS:
lrmod<-function(){
# Priors
# Note: Gelman recommends
# - scaling continuous predictors so they have mean 0 and sd = 0.5
# - using a non-informative Cauchy prior dt(0, pow(2.5,-2), 1)
# see arxiv.org/pdf/0901.4011.pdf
alpha ~ dt(0, pow(2.5, -2), 1)
beta ~ dt(0, pow(2.5, -2), 1)
# Likelihood
for(i in 1:n){
observed[i] ~ dbin(p[i], 1)
logitp[i] <- alpha + beta * voc[i]
p[i] <- exp(logitp[i]) / (1 + exp(logitp[i]))
# GOF test
presi[i] <- (observed[i] - p[i]) / sqrt(p[i] * (1 - p[i]))
obs.new[i] ~ dbin(p[i], 1)
presi.new[i] <- (obs.new[i] - p[i]) / sqrt(p[i] * (1 - p[i]))
D[i] <- pow(presi[i], 2)
D.new[i] <- pow(presi.new[i], 2)
}
fit <- sum(D[])
fit.new <- sum(D.new[])
}
# Bundle data
voc <- (exp.m$voc - mean(exp.m$voc)) / (0.5 * sd(exp.m$voc))
jagsdata <- list(observed = exp.m$observed, voc = voc, n = nrow(exp.m))
# Parameters to estimate
params <- c("alpha", "beta", "p", "presi", "fit", "fit.new")
# MCMC settings
nc <- 3
ni <- 3000
nb <- 1000
nt <- 2
out.p <- jags.parallel(data = jagsdata, parameters.to.save = params,
model.file = lrmod, n.thin= 2, n.chains = 3,
n.burnin = 1000, n.iter = 3000)
#Goodness-of-fit test
fitstats <- MCMCpstr(out.p, params = c("fit", "fit.new"), type = "chains")
T.extreme <- fitstats$fit.new >= fitstats$fit
(p.val <- mean(T.extreme))[1] 0.4546667Looking at the goodness-of-fit test (above), we again fail to reject the null hypothesis that our data are consisten with the assumed model. Below, we compare estimates of coefficients and 95% credible intervals to those obtained usin glm with scaled voc:
MCMCsummary(out.p, params = c("alpha", "beta"), round = 3) mean sd 2.5% 50% 97.5% Rhat n.eff
alpha -0.103 0.203 -0.487 -0.106 0.294 1 3068
beta -0.494 0.108 -0.708 -0.491 -0.286 1 3000exp.m$voc.scaled.gelman <- voc # scaled version
mod1b <- glm(exp.m$observed ~ voc.scaled.gelman, family = binomial(),
data = exp.m)
cbind(coef(mod1b), confint(mod1b))Waiting for profiling to be done... 2.5 % 97.5 %
(Intercept) -0.1045001 -0.4980208 0.2864257
voc.scaled.gelman -0.4870714 -0.7114791 -0.2835381What if we had naively used extremely vague priors for our regression parameters? We can compare the results using the MCMCplot function in the MCMCvis package (Figure 16.20).
lrmodv<-function(){
alpha~dnorm(0, 0.0001)
beta~dnorm(0, 0.0001)
# Likelihood
for(i in 1:n){
observed[i]~dbin(p[i],1)
logitp[i]<-alpha+beta*voc[i]
p[i]<-exp(logitp[i])/(1+exp(logitp[i]))
}
}
params <- c("alpha", "beta")
out.p.vague <- jags.parallel(data = jagsdata, parameters.to.save = params,
model.file = lrmodv, n.thin= 2, n.chains = 3,
n.burnin = 1000, n.iter = 3000)MCMCplot(object = out.p, object2=out.p.vague, params=c("alpha", "beta"),
offset=0.1, main='Posterior Distributions')
MCMCplot function in the MCMCvis package.
In this case, the priors make little difference to our end results, but that may not always be the case.
16.8 Aside: Logistic regression with multiple trials
If we have multiple observations for each unique set of predictor variables, then we can write the model as:
This formulation can provide significant increases in speed when fitting large data sets (see e.g., Iannarilli et al. 2019). The glm function allows us to fit a logistic regression model to grouped data if we specify the number of trials and the number of successes. We will demonstrate the approach using a famous data set from Bliss (1935), which contains the number of adult flour beetles killed after 5 hours of exposure to gaseous carbon disulfide at various concentrations. The data are contained in a number of different R packages, including the glmx package. We access the data using:
library(glmx)
data("BeetleMortality")
BeetleMortality dose died n
1 1.6907 6 59
2 1.7242 13 60
3 1.7552 18 62
4 1.7842 28 56
5 1.8113 52 63
6 1.8369 53 59
7 1.8610 61 62
8 1.8839 60 60Let
We can fit the model using:
lrdose <- glm(cbind(died, n - died) ~ dose, data = BeetleMortality, family = binomial())
summary(lrdose)
Call:
glm(formula = cbind(died, n - died) ~ dose, family = binomial(),
data = BeetleMortality)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -60.717 5.181 -11.72 <2e-16 ***
dose 34.270 2.912 11.77 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 284.202 on 7 degrees of freedom
Residual deviance: 11.232 on 6 degrees of freedom
AIC: 41.43
Number of Fisher Scoring iterations: 4To demonstrate that we get the same result if we fit a model to data containing a separate record for each observation, we create a “long format” data set below:
BeetleLong <- NULL
uid <- unique(BeetleMortality$dose)
for(i in seq_along(uid)){
tempdat <- BeetleMortality[i,]
BeetleLong <- rbind(BeetleLong,
data.frame(died = c(rep(1, tempdat$died), rep(0, tempdat$n- tempdat$died)),
dose = rep(tempdat$dose, tempdat$n)))
}
head(BeetleLong) died dose
1 1 1.6907
2 1 1.6907
3 1 1.6907
4 1 1.6907
5 1 1.6907
6 1 1.6907#Show we get the same group summaries as the original data set
BeetleLong %>% group_by(dose) %>%
dplyr::summarize(died = sum(died), trials = n())# A tibble: 8 × 3
dose died trials
<dbl> <dbl> <int>
1 1.69 6 59
2 1.72 13 60
3 1.76 18 62
4 1.78 28 56
5 1.81 52 63
6 1.84 53 59
7 1.86 61 62
8 1.88 60 60We then fit the same regression model to the set of Bernoulli trials showing that we get the same result:
lrdose2 <- glm(died ~ dose, data = BeetleLong, family = binomial())
summary(lrdose2)
Call:
glm(formula = died ~ dose, family = binomial(), data = BeetleLong)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -60.717 5.181 -11.72 <2e-16 ***
dose 34.270 2.912 11.77 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 645.44 on 480 degrees of freedom
Residual deviance: 372.47 on 479 degrees of freedom
AIC: 376.47
Number of Fisher Scoring iterations: 516.9 Aside: Complete separation
Occasionally, when fitting a logistic regression model, you may encounter a warning message that the glm fitting algorithm “did not converge” or that “fitted probabilities numerically 0 or 1 occurred.” This is indicative of a problem referred to as complete separation – in which a predictor variable or set of predictor variables is able to fully discriminate between the 0’s and 1’s.
To demonstrate, we will use an example developed by Jack Weiss using data from Piegorsch and Bailer (2005) collected to evaluate water quality and potential contamination by an industrial solvent trichloroethylene (TCE). The data are included in the Data4Ecologists package and can be accessed using:
library(Data4Ecologists)
Attaching package: 'Data4Ecologists'The following object is masked from 'package:ggeffects':
fishdata(wells)We will build a logistic regression model to describe the probability that a well will be contaminated with TCE as a function of the surrounding land use category (land.use) and whether or not sewers were used in the area surrounding the well (sewer). Each row in the data set contains a unique set of covariates, with y quantifying the number of wells contaminated with TSE and n giving the total number of wells with the same land.use and sewer covariates. Thus, we model the data using glm with a cbind(successes, failures) syntax as described in Section 16.8:
lr1 <- glm(cbind(y, n - y) ~ land.use + sewer, data = wells, family = binomial())
summary(lr1)
Call:
glm(formula = cbind(y, n - y) ~ land.use + sewer, family = binomial(),
data = wells)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.6568 0.7376 -4.957 7.14e-07 ***
land.usecomm 1.8676 0.8044 2.322 0.02025 *
land.useindus 2.2070 0.8053 2.741 0.00613 **
land.useinst 0.7584 0.8395 0.903 0.36629
land.userecr 0.6676 0.8435 0.791 0.42870
land.useresH 1.7316 0.7784 2.225 0.02611 *
land.useresL 0.6663 1.0501 0.635 0.52572
land.useresM 1.0212 0.7809 1.308 0.19099
land.usetrans 0.7933 0.8360 0.949 0.34267
land.useundev -18.3414 3033.9308 -0.006 0.99518
seweryes 1.5980 0.2955 5.407 6.41e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 146.956 on 19 degrees of freedom
Residual deviance: 15.201 on 9 degrees of freedom
AIC: 82.476
Number of Fisher Scoring iterations: 17Although we didn’t receive a warning in this case, we see the symptoms of complete separation if we inspect the model coefficients and their SEs; namely the coefficient for land.useundev is extremely large in absolute value (-18.34) and its SE is also large (3034).
Think-Pair-Share: Estimate the probability that a well placed in undeveloped land without sewers has TSE contamination. What do you think this indicates about the underlying data?
We can estimate the probability that a well placed in undeveloped land has TSE contamination by adding the intercept and the coefficient for land.useundev and then taking the inverse logit transformation:
betas.lr1 <- coef(lr1)
plogis(betas.lr1[1] + betas.lr1[length(betas.lr1)-1]) (Intercept)
2.794265e-10 We see that this probability is extremely small. Let’s look at the observations broken down by land use category:
wells %>% mutate(nocontam = n-y) %>% group_by(land.use) %>%
dplyr::summarize(contam = sum(y), noncontam = sum(nocontam))# A tibble: 10 × 3
land.use contam noncontam
<chr> <int> <int>
1 agri 2 53
2 comm 20 29
3 indus 20 25
4 inst 8 46
5 recr 7 57
6 resH 34 59
7 resL 2 24
8 resM 17 112
9 trans 8 51
10 undev 0 76We see that there are no wells in undeveloped areas that were contaminated. Thus, the smaller the coefficient for land.useundev the better – essentially, the numerical optimizer keeps moving towards
There are several different approaches that we might take to deal with this problem. One simple solution would be to group land use categories so that we no longer have complete separation. Another popular approach is to use penalized likelihood or Bayesian approaches (Firth 1993; Fijorek and Sokolowski 2012). Let’s consider a Bayesian model with
lrmod<-function(){
# Priors
for(i in 1:nlanduse){
beta.landuse[i] ~ dnorm(0, 1/3)
}
beta.sewer ~ dnorm(0, 1/3)
# Likelihood
for(i in 1:nobs){
y[i]~dbin(p[i], n[i])
logit(p[i]) <-beta.landuse[landuse[i]] + beta.sewer * sewer[i]
}
}
# Bundle data
jags.data <- list(y = wells$y,
landuse = as.numeric(as.factor(wells$land.use)),
sewer = as.numeric(as.factor(wells$sewer)),
n = wells$n, nobs = nrow(wells),
nlanduse = length(unique(wells$land.use)))
# Parameters to estimate
params <- c("beta.landuse", "beta.sewer")
# MCMC settings
nc <- 3
ni <- 3000
nb <- 1000
nt <- 2
lr.bayes <- jags.parallel(data = jags.data, parameters.to.save = params,
model.file = lrmod, n.thin = 1, n.chains = 3,
n.burnin = 1000, n.iter = 5000) Let’s look to make sure everything converged and inspect the SE’s of the different coefficients:
MCMCvis::MCMCsummary(lr.bayes) mean sd 2.5% 50% 97.5% Rhat
beta.landuse[1] -3.5816658 0.6176467 -4.9347325 -3.5403297 -2.4724330 1.02
beta.landuse[2] -1.4576318 0.4655273 -2.3580142 -1.4581147 -0.5485151 1.00
beta.landuse[3] -1.2347589 0.4630749 -2.1602480 -1.2348759 -0.3467499 1.00
beta.landuse[4] -2.6557703 0.4907499 -3.6626855 -2.6414482 -1.7253137 1.02
beta.landuse[5] -2.9142162 0.4867960 -3.9294930 -2.9049714 -2.0013045 1.00
beta.landuse[6] -1.6369983 0.4255751 -2.4760714 -1.6287131 -0.8162675 1.00
beta.landuse[7] -2.9165556 0.6427926 -4.2475171 -2.8842078 -1.7498260 1.00
beta.landuse[8] -2.6806829 0.3799699 -3.4104566 -2.6851236 -1.9373743 1.00
beta.landuse[9] -2.6881298 0.4667913 -3.6359707 -2.6831948 -1.7882780 1.00
beta.landuse[10] -4.8821682 0.8343893 -6.7384216 -4.8025198 -3.5205098 1.04
beta.sewer 0.6033508 0.2033223 0.2082786 0.6019894 0.9999361 1.00
deviance 86.2354926 7.0437185 74.1456128 85.6594947 101.6587751 1.00
n.eff
beta.landuse[1] 371
beta.landuse[2] 1445
beta.landuse[3] 1515
beta.landuse[4] 775
beta.landuse[5] 590
beta.landuse[6] 1552
beta.landuse[7] 577
beta.landuse[8] 901
beta.landuse[9] 850
beta.landuse[10] 154
beta.sewer 1281
deviance 1079The last landuse coefficient is the one for undeveloped. We see that the N(0, 3) prior helped to stabilize the posterior distribution for this coefficient. We can estimate the probability of a well in an undeveloped area without sewers being contaminated along with a 95% credible interval for this probability using:
# Estimate
MCMCvis::MCMCpstr(lr.bayes, params = "beta.landuse[10]", ISB = FALSE,
func = function(x){mean(plogis(x))})$`beta.landuse[10]`
[1] 0.01001391# 95% CI
MCMCvis::MCMCpstr(lr.bayes, params = "beta.landuse[10]", ISB = FALSE,
func = function(x){quantile(plogis(x), probs = c(0.025, 0.975))})$`beta.landuse[10]`
2.5% 97.5%
beta.landuse[10] 0.001183114 0.02873426The estimate is still very small, but non-zero. And, we are able to report an estimate of uncertainty to go with it.
The logistf package also provides a penalized likelihood approach developed by Firth (1993), which is equivalent to Bayesian logistic regression with a Jeffreys prior. The logistf function requires each observation to be binary (i.e., a Bernoulli response). Thus, we create a “long version” of our data set below.
library(logistf)
WellsLong <- NULL
for(i in 1:nrow(wells)){
tempdat <- wells[i,]
WellsLong <- rbind(WellsLong,
data.frame(contam = c(rep(1, tempdat$y), rep(0, tempdat$n- tempdat$y)),
land.use = rep(tempdat$land.use, tempdat$n),
sewer = rep(tempdat$sewer, tempdat$n)))
}Then, we fit the model using means coding so that we can easily compare our results to the Bayesian approach:
lrfirth <- logistf(contam ~ land.use + sewer -1, data = WellsLong, family=binomial())
summary(lrfirth)logistf(formula = contam ~ land.use + sewer - 1, data = WellsLong,
family = binomial())
Model fitted by Penalized ML
Coefficients:
coef se(coef) lower 0.95 upper 0.95 Chisq p
land.useagri -3.424344 0.6617365 -5.025055 -2.3136352 68.76325 1.110223e-16
land.usecomm -1.741409 0.3939727 -2.544436 -0.9788752 20.92229 4.782972e-06
land.useindus -1.409143 0.3872128 -2.200166 -0.6614146 14.05506 1.775357e-04
land.useinst -2.811149 0.4483773 -3.760190 -1.9761494 56.48525 5.662137e-14
land.userecr -2.895559 0.4450532 -3.853023 -2.0799366 68.76325 1.110223e-16
land.useresH -1.878656 0.3381481 -2.574947 -1.2278191 35.33950 2.769582e-09
land.useresL -2.770159 0.6920825 -4.415058 -1.5807259 28.47004 9.515975e-08
land.useresM -2.580059 0.3157102 -3.244995 -1.9922454 Inf 0.000000e+00
land.usetrans -2.777363 0.4372769 -3.707734 -1.9676855 60.79674 6.328271e-15
land.useundev -5.632506 1.4246979 -10.478750 -3.6571844 Inf 0.000000e+00
seweryes 1.555010 0.2842178 1.004490 2.1479905 33.46428 7.258474e-09
method
land.useagri 2
land.usecomm 2
land.useindus 2
land.useinst 2
land.userecr 2
land.useresH 2
land.useresL 2
land.useresM 2
land.usetrans 2
land.useundev 2
seweryes 2
Method: 1-Wald, 2-Profile penalized log-likelihood, 3-None
Likelihood ratio test=347.4569 on 11 df, p=0, n=650
Wald test = 168.1994 on 11 df, p = 0We see again that the estimate of the coefficient for the undeveloped category is shrunk back towards 0, but not as much as in our Bayesian model using the N(0, 3) prior.
16.10 References
The distribution of the
Here we use the
dstpfunction in theLaplacesDemonpackage (Statisticat and LLC. 2021) since it is parameterized in the same way as the dt function in JAGS↩︎